Classification and nomenclature system for human Alphapapillomavirus variants: general features, nucleotide landmarks and assignment of HPV6 and HPV11 isolates to variant lineages
- PMID: 22131111
- PMCID: PMC3690374
Classification and nomenclature system for human Alphapapillomavirus variants: general features, nucleotide landmarks and assignment of HPV6 and HPV11 isolates to variant lineages
Abstract
Background: Papillomaviruses constitute a family of viruses that can be classified into genera, species and types based on their viral genome heterogeneity. Currently circulating infectious human Alphapapillomaviruses (alpha-PVs) constitute a set of viral genomes that have evolved from archaic times and display features of host co-speciation. Viral variants are more recently evolved genomes that require a standardized classification and nomenclature.
Objectives: To describe a system for the classification and nomenclature of HPV viral variants and provide landmarks for the numbering of nucleotide positions.
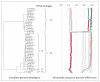
Methods: The complete 8 kb genomes of the alpha-9 species group and HPV6 and 11 types, collected from isolates throughout the world were obtained from published reports and GenBank. Complete genomes for each HPV type were aligned using the E1 start codon and sequence divergence was calculated by global and pairwise alignments using the MUSCLE program. Phylogenetic trees were constructed from the aligned sequences using a maximum likelihood method (RAxML).
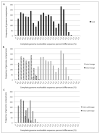
Results: Pairwise comparisons of nucleotide differences between complete genomes of each type from alpha-9 HPV isolates (HPV16, 31, 33, 35, 52, 58 and 67) revealed a trimodal distribution. Maximum heterogeneity for variants within a type varied from 0.6%-2.3%. Nucleotide differences of approximately 1.0%-10.0% and 0.5%-1.0% of the complete genomes were used to define variant lineages and sublineages, respectively. Analysis of 43 HPV6 complete genomes indicated the presence of 2 variant lineages, whereas 32 HPV11 isolates were highly similar and clustered into 2 sublineages. A table was constructed of the human alpha-PV landmark nucleotide sequences for future reference and alignments.
Conclusions: A proposed nomenclature system for viral variants and coordination of nucleotide positions will facilitate the comparison of variants across geographic regions and amongst different populations. In addition, this system will facilitate study of pathogenic, tissue tropism and functional differences amongst variant lineages of and polymorphisms within HPV variants.
Figures

References
-
- Herbst LH, Lenz J, Van Doorslaer K, Chen Z, Stacy BA, Wellehan JF, Manire CA, Burk RD. Genomic characterization of two novel reptilian papillomaviruses, Chelonia mydas papillomavirus 1 and Caretta caretta papillomavirus 1. Virology. 2009;383:131–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

