Detection of somatic copy number alterations in cancer using targeted exome capture sequencing
- PMID: 22131877
- PMCID: PMC3223606
- DOI: 10.1593/neo.111252
Detection of somatic copy number alterations in cancer using targeted exome capture sequencing
Abstract
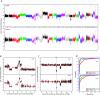
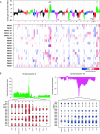
The research community at large is expending considerable resources to sequence the coding region of the genomes of tumors and other human diseases using targeted exome capture (i.e., "whole exome sequencing"). The primary goal of targeted exome sequencing is to identify nonsynonymous mutations that potentially have functional consequences. Here, we demonstrate that whole-exome sequencing data can also be analyzed for comprehensively monitoring somatic copy number alterations (CNAs) by benchmarking the technique against conventional array CGH. A series of 17 matched tumor and normal tissues from patients with metastatic castrate-resistant prostate cancer was used for this assessment. We show that targeted exome sequencing reliably identifies CNAs that are common in advanced prostate cancer, such as androgen receptor (AR) gain and PTEN loss. Taken together, these data suggest that targeted exome sequencing data can be effectively leveraged for the detection of somatic CNAs in cancer.
Figures
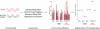
References
-
- Pinkel D, Seagraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. - PubMed
-
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Månér S, Massa H, Walker M, Chi M, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. - PubMed
-
- McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, deBakker PI, Maller JB, Kirby A, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
