DNA methylation profiles of primary colorectal carcinoma and matched liver metastasis
- PMID: 22132162
- PMCID: PMC3221680
- DOI: 10.1371/journal.pone.0027889
DNA methylation profiles of primary colorectal carcinoma and matched liver metastasis
Abstract
Background: The contribution of DNA methylation to the metastatic process in colorectal cancers (CRCs) is unclear.
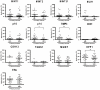
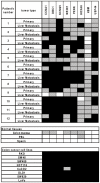
Methods: We evaluated the methylation status of 13 genes (MINT1, MINT2, MINT31, MLH1, p16, p14, TIMP3, CDH1, CDH13, THBS1, MGMT, HPP1 and ERα) by bisulfite-pyrosequencing in 79 CRCs comprising 36 CRCs without liver metastasis and 43 CRCs with liver metastasis, including 16 paired primary CRCs and liver metastasis. We also performed methylated CpG island amplification microarrays (MCAM) in three paired primary and metastatic cancers.
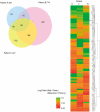
Results: Methylation of p14, TIMP3 and HPP1 in primary CRCs progressively decreased from absence to presence of liver metastasis (13.1% vs. 4.3%; 14.8% vs. 3.7%; 43.9% vs. 35.8%, respectively) (P<.05). When paired primary and metastatic tumors were compared, only MGMT methylation was significantly higher in metastatic cancers (27.4% vs. 13.4%, P = .013), and this difference was due to an increase in methylation density rather than frequency in the majority of cases. MCAM showed an average 7.4% increase in DNA methylated genes in the metastatic samples. The numbers of differentially hypermethylated genes in the liver metastases increased with increasing time between resection of the primary and resection of the liver metastasis. Bisulfite-pyrosequencing validation in 12 paired samples showed that most of these increases were not conserved, and could be explained by differences in methylation density rather than frequency.
Conclusions: Most DNA methylation differences between primary CRCs and matched liver metastasis are due to random variation and an increase in DNA methylation density rather than de-novo inactivation and silencing. Thus, DNA methylation changes occur for the most part before progression to liver metastasis.
Conflict of interest statement
Figures

Similar articles
-
Rare CpG island methylator phenotype in ulcerative colitis-associated neoplasias.Gastroenterology. 2007 Apr;132(4):1254-60. doi: 10.1053/j.gastro.2007.01.035. Epub 2007 Jan 25. Gastroenterology. 2007. PMID: 17408633
-
Distinct profiles of epigenetic evolution between colorectal cancers with and without metastasis.Am J Pathol. 2011 Apr;178(4):1835-46. doi: 10.1016/j.ajpath.2010.12.045. Epub 2011 Mar 4. Am J Pathol. 2011. PMID: 21406167 Free PMC article.
-
The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer.Gastroenterology. 2007 Jan;132(1):127-38. doi: 10.1053/j.gastro.2006.09.018. Epub 2006 Sep 20. Gastroenterology. 2007. PMID: 17087942
-
CpG island methylation of genes accumulates during the adenoma progression step of the multistep pathogenesis of colorectal cancer.Genes Chromosomes Cancer. 2006 Aug;45(8):781-9. doi: 10.1002/gcc.20341. Genes Chromosomes Cancer. 2006. PMID: 16708352
-
The DNA hypermethylation phenotype of colorectal cancer liver metastases resembles that of the primary colorectal cancers.BMC Cancer. 2020 Apr 6;20(1):290. doi: 10.1186/s12885-020-06777-6. BMC Cancer. 2020. PMID: 32252665 Free PMC article.
Cited by
-
Fusobacterium in colonic flora and molecular features of colorectal carcinoma.Cancer Res. 2014 Mar 1;74(5):1311-8. doi: 10.1158/0008-5472.CAN-13-1865. Epub 2014 Jan 2. Cancer Res. 2014. PMID: 24385213 Free PMC article.
-
Temozolomide in the Era of Precision Medicine.Cancer Res. 2017 Feb 15;77(4):823-826. doi: 10.1158/0008-5472.CAN-16-2983. Epub 2017 Feb 3. Cancer Res. 2017. PMID: 28159862 Free PMC article. Review.
-
The transition from primary colorectal cancer to isolated peritoneal malignancy is associated with an increased tumour mutational burden.Sci Rep. 2020 Nov 3;10(1):18900. doi: 10.1038/s41598-020-75844-6. Sci Rep. 2020. PMID: 33144643 Free PMC article.
-
Biological factors driving colorectal cancer metastasis.World J Gastrointest Oncol. 2024 Feb 15;16(2):259-272. doi: 10.4251/wjgo.v16.i2.259. World J Gastrointest Oncol. 2024. PMID: 38425391 Free PMC article. Review.
-
MGMT is frequently inactivated in pancreatic NET-G2 and is associated with the therapeutic activity of STZ-based regimens.Sci Rep. 2023 May 9;13(1):7535. doi: 10.1038/s41598-023-34666-y. Sci Rep. 2023. PMID: 37161026 Free PMC article.
References
-
- Jemal A, Siegel R, Ward E, Murray T, Xu J, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. - PubMed
-
- Potter JD, Slattery ML, Bostick RM, Gapstur SM. Colon cancer: a review of the epidemiology. Epidemiol Rev. 1993;15:499–545. - PubMed
-
- Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

