Genome-wide identification of microRNAs in response to low nitrate availability in maize leaves and roots
- PMID: 22132192
- PMCID: PMC3223196
- DOI: 10.1371/journal.pone.0028009
Genome-wide identification of microRNAs in response to low nitrate availability in maize leaves and roots
Abstract
Background: Nitrate is the major source of nitrogen available for many crop plants and is often the limiting factor for plant growth and agricultural productivity especially for maize. Many studies have been done identifying the transcriptome changes under low nitrate conditions. However, the microRNAs (miRNAs) varied under nitrate limiting conditions in maize has not been reported. MiRNAs play important roles in abiotic stress responses and nutrient deprivation.
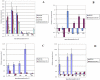
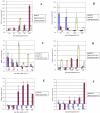
Methodology/principal findings: In this study, we used the SmartArray™ and GeneChip® microarray systems to perform a genome-wide search to detect miRNAs responding to the chronic and transient nitrate limiting conditions in maize. Nine miRNA families (miR164, miR169, miR172, miR397, miR398, miR399, miR408, miR528, and miR827) were identified in leaves, and nine miRNA families (miR160, miR167, miR168, miR169, miR319, miR395, miR399, miR408, and miR528) identified in roots. They were verified by real time stem loop RT-PCR, and some with additional time points of nitrate limitation. The miRNAs identified showed overlapping or unique responses to chronic and transient nitrate limitation, as well as tissue specificity. The potential target genes of these miRNAs in maize were identified. The expression of some of these was examined by qRT-PCR. The potential function of these miRNAs in responding to nitrate limitation is described.
Conclusions/significance: Genome-wide miRNAs responding to nitrate limiting conditions in maize leaves and roots were identified. This provides an insight into the timing and tissue specificity of the transcriptional regulation to low nitrate availability in maize. The knowledge gained will help understand the important roles miRNAs play in maize responding to a nitrogen limiting environment and eventually develop strategies for the improvement of maize genetics.
Conflict of interest statement
Figures

References
-
- Guo JH, Liu XJ, Zhang Y, Shen JL, Han WX, et al. Significant Acidification in Major Chinese Croplands. Science. 2010;327:1008–1010. - PubMed
-
- Tilman D, Cassman KG, Matson PA, Naylor R, Polasky S. Agricultural sustainability and intensive production practices. Nature. 2002;418:671–677. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

