Microbial biogeography of public restroom surfaces
- PMID: 22132229
- PMCID: PMC3223236
- DOI: 10.1371/journal.pone.0028132
Microbial biogeography of public restroom surfaces
Abstract
We spend the majority of our lives indoors where we are constantly exposed to bacteria residing on surfaces. However, the diversity of these surface-associated communities is largely unknown. We explored the biogeographical patterns exhibited by bacteria across ten surfaces within each of twelve public restrooms. Using high-throughput barcoded pyrosequencing of the 16 S rRNA gene, we identified 19 bacterial phyla across all surfaces. Most sequences belonged to four phyla: Actinobacteria, Bacteriodetes, Firmicutes and Proteobacteria. The communities clustered into three general categories: those found on surfaces associated with toilets, those on the restroom floor, and those found on surfaces routinely touched with hands. On toilet surfaces, gut-associated taxa were more prevalent, suggesting fecal contamination of these surfaces. Floor surfaces were the most diverse of all communities and contained several taxa commonly found in soils. Skin-associated bacteria, especially the Propionibacteriaceae, dominated surfaces routinely touched with our hands. Certain taxa were more common in female than in male restrooms as vagina-associated Lactobacillaceae were widely distributed in female restrooms, likely from urine contamination. Use of the SourceTracker algorithm confirmed many of our taxonomic observations as human skin was the primary source of bacteria on restroom surfaces. Overall, these results demonstrate that restroom surfaces host relatively diverse microbial communities dominated by human-associated bacteria with clear linkages between communities on or in different body sites and those communities found on restroom surfaces. More generally, this work is relevant to the public health field as we show that human-associated microbes are commonly found on restroom surfaces suggesting that bacterial pathogens could readily be transmitted between individuals by the touching of surfaces. Furthermore, we demonstrate that we can use high-throughput analyses of bacterial communities to determine sources of bacteria on indoor surfaces, an approach which could be used to track pathogen transmission and test the efficacy of hygiene practices.
Conflict of interest statement
Figures
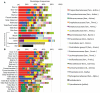


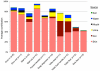
References
-
- Ojima M, Toshima Y, Koya E, Ara K, Tokuda H, et al. Hygiene measures considering actual distributions of microorganisms in Japanese households. Journal of Applied Microbiology. 2002;93:800–809. - PubMed
-
- Medrano-Felix A, Martinez C, Castro-del Campo N, Leon-Felix J, Peraza-Garay F, et al. Impact of prescribed cleaning and disinfectant use on microbial contamination in the home. Journal of Applied Microbiology. 2011;110:463–471. - PubMed
-
- Ojima M, Toshima Y, Koya E, Ara K, Kawai S, et al. Bacterial contamination of Japanese households and related concern about sanitation. International Journal of Environmental Health Research. 2002;12:41–52. - PubMed
-
- Rusin P, Orosz-Coughlin P, Gerba C. Reduction of faecal coliform, coliform and heterotrophic plate count bacteria in the household kitchen and bathroom by disinfection with hypochlorite cleaners. Journal of Applied Microbiology. 1998;85:819–828. - PubMed

