Self-organizing circuit assembly through spatiotemporally coordinated neuronal migration within geometric constraints
- PMID: 22132234
- PMCID: PMC3222678
- DOI: 10.1371/journal.pone.0028156
Self-organizing circuit assembly through spatiotemporally coordinated neuronal migration within geometric constraints
Abstract
Background: Neurons are dynamically coupled with each other through neurite-mediated adhesion during development. Understanding the collective behavior of neurons in circuits is important for understanding neural development. While a number of genetic and activity-dependent factors regulating neuronal migration have been discovered on single cell level, systematic study of collective neuronal migration has been lacking. Various biological systems are shown to be self-organized, and it is not known if neural circuit assembly is self-organized. Besides, many of the molecular factors take effect through spatial patterns, and coupled biological systems exhibit emergent property in response to geometric constraints. How geometric constraints of the patterns regulate neuronal migration and circuit assembly of neurons within the patterns remains unexplored.
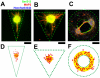
Methodology/principal findings: We established a two-dimensional model for studying collective neuronal migration of a circuit, with hippocampal neurons from embryonic rats on Matrigel-coated self-assembled monolayers (SAMs). When the neural circuit is subject to geometric constraints of a critical scale, we found that the collective behavior of neuronal migration is spatiotemporally coordinated. Neuronal somata that are evenly distributed upon adhesion tend to aggregate at the geometric center of the circuit, forming mono-clusters. Clustering formation is geometry-dependent, within a critical scale from 200 µm to approximately 500 µm. Finally, somata clustering is neuron-type specific, and glutamatergic and GABAergic neurons tend to aggregate homo-philically.
Conclusions/significance: We demonstrate self-organization of neural circuits in response to geometric constraints through spatiotemporally coordinated neuronal migration, possibly via mechanical coupling. We found that such collective neuronal migration leads to somata clustering, and mono-cluster appears when the geometric constraints fall within a critical scale. The discovery of geometry-dependent collective neuronal migration and the formation of somata clustering in vitro shed light on neural development in vivo.
Conflict of interest statement
Figures






Similar articles
-
Diminished neuronal activity increases neuron-neuron connectivity underlying silent synapse formation and the rapid conversion of silent to functional synapses.J Neurosci. 2005 Apr 20;25(16):4040-51. doi: 10.1523/JNEUROSCI.4115-04.2005. J Neurosci. 2005. PMID: 15843606 Free PMC article.
-
Innate synchronous oscillations in freely-organized small neuronal circuits.PLoS One. 2010 Dec 28;5(12):e14443. doi: 10.1371/journal.pone.0014443. PLoS One. 2010. PMID: 21203438 Free PMC article.
-
Surface coating as a key parameter in engineering neuronal network structures in vitro.Biointerphases. 2012 Dec;7(1-4):29. doi: 10.1007/s13758-012-0029-7. Epub 2012 Apr 24. Biointerphases. 2012. PMID: 22589072
-
Synapse formation in developing neural circuits.Curr Top Dev Biol. 2009;87:53-79. doi: 10.1016/S0070-2153(09)01202-2. Curr Top Dev Biol. 2009. PMID: 19427516 Free PMC article. Review.
-
Role of mechanical cues in shaping neuronal morphology and connectivity.Biol Cell. 2018 Jun;110(6):125-136. doi: 10.1111/boc.201800003. Epub 2018 May 14. Biol Cell. 2018. PMID: 29698566 Review.
Cited by
-
Tracking axon initial segment plasticity using high-density microelectrode arrays: A computational study.Front Neuroinform. 2022 Oct 3;16:957255. doi: 10.3389/fninf.2022.957255. Front Neuroinform. 2022. PMID: 36221258 Free PMC article.
-
Self-organization of modular network architecture by activity-dependent neuronal migration and outgrowth.Elife. 2019 Sep 17;8:e47996. doi: 10.7554/eLife.47996. Elife. 2019. PMID: 31526478 Free PMC article.
-
Emergence of assortative mixing between clusters of cultured neurons.PLoS Comput Biol. 2014 Sep 4;10(9):e1003796. doi: 10.1371/journal.pcbi.1003796. eCollection 2014 Sep. PLoS Comput Biol. 2014. PMID: 25188377 Free PMC article.
-
Galvanotactic control of collective cell migration in epithelial monolayers.Nat Mater. 2014 Apr;13(4):409-17. doi: 10.1038/nmat3891. Epub 2014 Mar 9. Nat Mater. 2014. PMID: 24608142
-
Small-world networks of neuroblastoma cells cultured in three-dimensional polymeric scaffolds featuring multi-scale roughness.Neural Regen Res. 2020 Apr;15(4):759-768. doi: 10.4103/1673-5374.266923. Neural Regen Res. 2020. PMID: 31638101 Free PMC article.
References
-
- Ayala R, Shu TZ, Tsai LH. Trekking across the brain: The journey of neuronal migration. Cell. 2007;128:29–43. - PubMed
-
- Kriegstein AR, Noctor SC. Patterns of neuronal migration in the embryonic cortex. Trends in Neurosciences. 2004;27:392–399. - PubMed
-
- Marin O, Rubenstein JLR. Cell migration in the forebrain. Annual Review of Neuroscience. 2003;26:441–483. - PubMed
-
- Gupta A, Tsai LH, Wynshaw-Boris A. Life is a journey: A genetic look at neocortical development. Nature Reviews Genetics. 2002;3:342–355. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

