Assembly and proteolytic processing of mycobacterial ClpP1 and ClpP2
- PMID: 22132756
- PMCID: PMC3258218
- DOI: 10.1186/1471-2091-12-61
Assembly and proteolytic processing of mycobacterial ClpP1 and ClpP2
Abstract
Background: Caseinolytic proteases (ClpPs) are barrel-shaped self-compartmentalized peptidases involved in eliminating damaged or short-lived regulatory proteins. The Mycobacterium tuberculosis (MTB) genome contains two genes coding for putative ClpPs, ClpP1 and ClpP2 respectively, that are likely to play a role in the virulence of the bacterium.
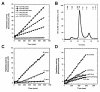
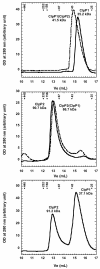
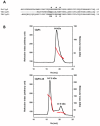
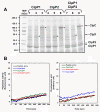
Results: We report the first biochemical characterization of ClpP1 and ClpP2 peptidases from MTB. Both proteins were produced and purified in Escherichia coli. Use of fluorogenic model peptides of diverse specificities failed to show peptidase activity with recombinant mycobacterial ClpP1 or ClpP2. However, we found that ClpP1 had a proteolytic activity responsible for its own cleavage after the Arg8 residue and cleavage of ClpP2 after the Ala12 residue. In addition, we showed that the absence of any peptidase activity toward model peptides was not due to an obstruction of the entry pore by the N-terminal flexible extremity of the proteins, nor to an absolute requirement for the ClpX or ClpC ATPase complex. Finally, we also found that removing the putative propeptides of ClpP1 and ClpP2 did not result in cleavage of model peptides. We have also shown that recombinant ClpP1 and ClpP2 do not assemble in the conventional functional tetradecameric form but in lower order oligomeric species ranging from monomers to heptamers. The concomitant presence of both ClpP1 and ClpP2 did not result in tetradecameric assembly. Deleting the amino-terminal extremity of ClpP1 and ClpP2 (the putative propeptide or entry gate) promoted the assembly in higher order oligomeric species, suggesting that the flexible N-terminal extremity of mycobacterial ClpPs participated in the destabilization of interaction between heptamers.
Conclusion: Despite the conservation of a Ser protease catalytic triad in their primary sequences, mycobacterial ClpP1 and ClpP2 do not have conventional peptidase activity toward peptide models and display an unusual mechanism of self-assembly. Therefore, the mechanism underlying their peptidase and proteolytic activities might differ from that of other ClpP proteolytic complexes.
Figures



References
-
- Gottesman S. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol. 2003;19:565–587. - PubMed
-
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426(6968):895–899. - PubMed
-
- Wang J, Hartling JA, Flanagan JM. The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell. 1997;91(4):447–456. - PubMed
-
- Thompson MW, Maurizi MR. Activity and specificity of Escherichia coli ClpAP protease in cleaving model peptide substrates. J Biol Chem. 1994;269(27):18201–18208. - PubMed
-
- Woo KM, Chung WJ, Ha DB, Goldberg AL, Chung CH. Protease Ti from Escherichia coli requires ATP hydrolysis for protein breakdown but not for hydrolysis of small peptides. J Biol Chem. 1989;264(4):2088–2091. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

