Revealing the missing expressed genes beyond the human reference genome by RNA-Seq
- PMID: 22133125
- PMCID: PMC3288009
- DOI: 10.1186/1471-2164-12-590
Revealing the missing expressed genes beyond the human reference genome by RNA-Seq
Abstract
Background: The complete and accurate human reference genome is important for functional genomics researches. Therefore, the incomplete reference genome and individual specific sequences have significant effects on various studies.
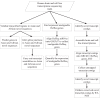
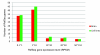
Results: we used two RNA-Seq datasets from human brain tissues and 10 mixed cell lines to investigate the completeness of human reference genome. First, we demonstrated that in previously identified ~5 Mb Asian and ~5 Mb African novel sequences that are absent from the human reference genome of NCBI build 36, ~211 kb and ~201 kb of them could be transcribed, respectively. Our results suggest that many of those transcribed regions are not specific to Asian and African, but also present in Caucasian. Then, we found that the expressions of 104 RefSeq genes that are unalignable to NCBI build 37 in brain and cell lines are higher than 0.1 RPKM. 55 of them are conserved across human, chimpanzee and macaque, suggesting that there are still a significant number of functional human genes absent from the human reference genome. Moreover, we identified hundreds of novel transcript contigs that cannot be aligned to NCBI build 37, RefSeq genes and EST sequences. Some of those novel transcript contigs are also conserved among human, chimpanzee and macaque. By positioning those contigs onto the human genome, we identified several large deletions in the reference genome. Several conserved novel transcript contigs were further validated by RT-PCR.
Conclusion: Our findings demonstrate that a significant number of genes are still absent from the incomplete human reference genome, highlighting the importance of further refining the human reference genome and curating those missing genes. Our study also shows the importance of de novo transcriptome assembly. The comparative approach between reference genome and other related human genomes based on the transcriptome provides an alternative way to refine the human reference genome.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

