Visual detection of gene mutations based on isothermal strand-displacement polymerase reaction and lateral flow strip
- PMID: 22133519
- PMCID: PMC3248977
- DOI: 10.1016/j.bios.2011.10.037
Visual detection of gene mutations based on isothermal strand-displacement polymerase reaction and lateral flow strip
Abstract
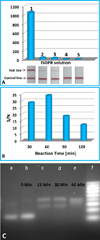
Here, we describe a simple and sensitive approach for visual detection of gene mutations based on isothermal strand-displacement polymerase reactions (ISDPR) and lateral flow strip (LFS). The concept was first demonstrated by detecting the R156H-mutant gene of keratin 10 in Epidermolytic hyperkeratosis (EHK). In the presence of biotin-modified hairpin DNA and digoxin-modified primer, the R156H-mutant DNA triggered the ISDPR to produce numerous digoxin- and biotin-attached duplex DNA products. The product was detected on the LFS through dual immunoreactions (anti-digoxin antibody on the gold nanoparticle (Au-NP) and digoxin on the duplex, anti-biotin antibody on the LFS test zone and biotin on the duplex). The accumulation of Au-NPs produced the characteristic red band, enabling visual detection of the mutant gene without instrumentation. After systematic optimization of the ISDPR experimental conditions and the parameters of the assay, the current approach was capable of detecting as low as 1-fM R156H-mutant DNA within 75 min without instrumentation. Differentiation of R156H- and R156C-mutant DNA on the R156 mutation site was realized by using fluorescein- and biotin-modified hairpin probes in the ISDPR process. The approach thus provides a simple, sensitive, and low-cost tool for the detection of gene mutations.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures




References
-
- Brown PO, Botstein D. Nature Genet. 1999;21:33–37. - PubMed
-
- Gill P, Ghaemi A. Nucleosides, Nucleotides, and Nucleic Acids. 2008;27:224–243. - PubMed
-
- Haruna K, Suga Y, Mizuno Y, Hasegawa T, Kourou K, Matsuba S, Muramatsu S, Ikeda S. J. Dermatology. 2007;34:545–548. - PubMed
-
- He Y, Zeng K, Zhang X, Gurung A, Baloda M, Xu H, Liu G. Electrochem. Commun. 2010a;12(7):985–988.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

