Promiscuous DNA synthesis by human DNA polymerase θ
- PMID: 22135286
- PMCID: PMC3315306
- DOI: 10.1093/nar/gkr1102
Promiscuous DNA synthesis by human DNA polymerase θ
Abstract
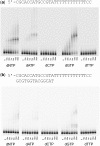
The biological role of human DNA polymerase θ (POLQ) is not yet clearly defined, but it has been proposed to participate in several cellular processes based on its translesion synthesis capabilities. POLQ is a low-fidelity polymerase capable of efficient bypass of blocking lesions such as abasic sites and thymine glycols as well as extension of mismatched primer termini. Here, we show that POLQ possesses a DNA polymerase activity that appears to be template independent and allows efficient extension of single-stranded DNA as well as duplex DNA with either protruding or multiply mismatched 3'-OH termini. We hypothesize that this DNA synthesis activity is related to the proposed role for POLQ in the repair or tolerance of double-strand breaks.
Figures





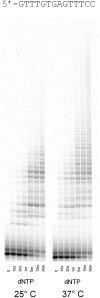
Similar articles
-
DNA polymerase θ-mediated repair of high LET radiation-induced complex DNA double-strand breaks.Nucleic Acids Res. 2023 Mar 21;51(5):2257-2269. doi: 10.1093/nar/gkad076. Nucleic Acids Res. 2023. PMID: 36805268 Free PMC article.
-
DNA polymerase theta (POLQ) can extend from mismatches and from bases opposite a (6-4) photoproduct.DNA Repair (Amst). 2008 Jan 1;7(1):119-27. doi: 10.1016/j.dnarep.2007.08.005. Epub 2007 Oct 24. DNA Repair (Amst). 2008. PMID: 17920341 Free PMC article.
-
Promiscuous mismatch extension by human DNA polymerase lambda.Nucleic Acids Res. 2006 Jun 28;34(11):3259-66. doi: 10.1093/nar/gkl377. Print 2006. Nucleic Acids Res. 2006. PMID: 16807316 Free PMC article.
-
Targeting the DNA Repair Enzyme Polymerase θ in Cancer Therapy.Trends Cancer. 2021 Feb;7(2):98-111. doi: 10.1016/j.trecan.2020.09.007. Epub 2020 Oct 24. Trends Cancer. 2021. PMID: 33109489 Review.
-
Biochemical basis of DNA replication fidelity.Crit Rev Biochem Mol Biol. 1993;28(2):83-126. doi: 10.3109/10409239309086792. Crit Rev Biochem Mol Biol. 1993. PMID: 8485987 Review.
Cited by
-
One-step enzymatic modification of RNA 3' termini using polymerase θ.Nucleic Acids Res. 2019 Apr 23;47(7):3272-3283. doi: 10.1093/nar/gkz029. Nucleic Acids Res. 2019. PMID: 30818397 Free PMC article.
-
How to use CRISPR/Cas9 in plants: from target site selection to DNA repair.J Exp Bot. 2024 Sep 11;75(17):5325-5343. doi: 10.1093/jxb/erae147. J Exp Bot. 2024. PMID: 38648173 Free PMC article. Review.
-
Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway?Trends Biochem Sci. 2015 Nov;40(11):701-714. doi: 10.1016/j.tibs.2015.08.006. Epub 2015 Oct 1. Trends Biochem Sci. 2015. PMID: 26439531 Free PMC article. Review.
-
The helicase domain of Polθ counteracts RPA to promote alt-NHEJ.Nat Struct Mol Biol. 2017 Dec;24(12):1116-1123. doi: 10.1038/nsmb.3494. Epub 2017 Oct 23. Nat Struct Mol Biol. 2017. PMID: 29058711 Free PMC article.
-
Reclassification of family A DNA polymerases reveals novel functional subfamilies and distinctive structural features.Nucleic Acids Res. 2023 May 22;51(9):4488-4507. doi: 10.1093/nar/gkad242. Nucleic Acids Res. 2023. PMID: 37070157 Free PMC article.
References
-
- Bebenek K, Joyce CM, Fitzgerald MP, Kunkel TA. The fidelity of DNA synthesis catalyzed by derivatives of Escherichia coli DNA polymerase I. J. Biol. Chem. 1990;265:13878–13887. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

