NONCODE v3.0: integrative annotation of long noncoding RNAs
- PMID: 22135294
- PMCID: PMC3245065
- DOI: 10.1093/nar/gkr1175
NONCODE v3.0: integrative annotation of long noncoding RNAs
Abstract
Facilitated by the rapid progress of high-throughput sequencing technology, a large number of long noncoding RNAs (lncRNAs) have been identified in mammalian transcriptomes over the past few years. LncRNAs have been shown to play key roles in various biological processes such as imprinting control, circuitry controlling pluripotency and differentiation, immune responses and chromosome dynamics. Notably, a growing number of lncRNAs have been implicated in disease etiology. With the increasing number of published lncRNA studies, the experimental data on lncRNAs (e.g. expression profiles, molecular features and biological functions) have accumulated rapidly. In order to enable a systematic compilation and integration of this information, we have updated the NONCODE database (http://www.noncode.org) to version 3.0 to include the first integrated collection of expression and functional lncRNA data obtained from re-annotated microarray studies in a single database. NONCODE has a user-friendly interface with a variety of search or browse options, a local Genome Browser for visualization and a BLAST server for sequence-alignment search. In addition, NONCODE provides a platform for the ongoing collation of ncRNAs reported in the literature. All data in NONCODE are open to users, and can be downloaded through the website or obtained through the SOAP API and DAS services.
Figures
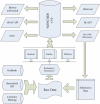
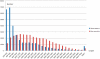
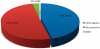
Similar articles
-
NONCODE v2.0: decoding the non-coding.Nucleic Acids Res. 2008 Jan;36(Database issue):D170-2. doi: 10.1093/nar/gkm1011. Epub 2007 Nov 13. Nucleic Acids Res. 2008. PMID: 18000000 Free PMC article.
-
NONCODEv4: exploring the world of long non-coding RNA genes.Nucleic Acids Res. 2014 Jan;42(Database issue):D98-103. doi: 10.1093/nar/gkt1222. Epub 2013 Nov 26. Nucleic Acids Res. 2014. PMID: 24285305 Free PMC article.
-
NONCODE: an integrated knowledge database of non-coding RNAs.Nucleic Acids Res. 2005 Jan 1;33(Database issue):D112-5. doi: 10.1093/nar/gki041. Nucleic Acids Res. 2005. PMID: 15608158 Free PMC article.
-
Review: Long non-coding RNA in livestock.Animal. 2020 Oct;14(10):2003-2013. doi: 10.1017/S1751731120000841. Epub 2020 May 8. Animal. 2020. PMID: 32381139 Review.
-
Computational analysis of noncoding RNAs.Wiley Interdiscip Rev RNA. 2012 Nov-Dec;3(6):759-78. doi: 10.1002/wrna.1134. Epub 2012 Sep 18. Wiley Interdiscip Rev RNA. 2012. PMID: 22991327 Free PMC article. Review.
Cited by
-
PIWI-Interacting RNAs: A Pivotal Regulator in Neurological Development and Disease.Genes (Basel). 2024 May 21;15(6):653. doi: 10.3390/genes15060653. Genes (Basel). 2024. PMID: 38927589 Free PMC article. Review.
-
Long non-coding RNA Databases in Cardiovascular Research.Genomics Proteomics Bioinformatics. 2016 Aug;14(4):191-9. doi: 10.1016/j.gpb.2016.03.001. Epub 2016 Apr 2. Genomics Proteomics Bioinformatics. 2016. PMID: 27049585 Free PMC article. Review.
-
Identification of Stably Expressed lncRNAs as Valid Endogenous Controls for Profiling of Human Glioma.J Cancer. 2015 Jan 1;6(2):111-9. doi: 10.7150/jca.10867. eCollection 2015. J Cancer. 2015. PMID: 25561975 Free PMC article.
-
Online tools for bioinformatics analyses in nutrition sciences.Adv Nutr. 2012 Sep 1;3(5):654-65. doi: 10.3945/an.112.002477. Adv Nutr. 2012. PMID: 22983844 Free PMC article. Review.
-
Accurate Prediction of Transposon-Derived piRNAs by Integrating Various Sequential and Physicochemical Features.PLoS One. 2016 Apr 13;11(4):e0153268. doi: 10.1371/journal.pone.0153268. eCollection 2016. PLoS One. 2016. PMID: 27074043 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

