PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse
- PMID: 22135298
- PMCID: PMC3245126
- DOI: 10.1093/nar/gkr1122
PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse
Abstract
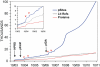
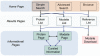
PhosphoSitePlus (http://www.phosphosite.org) is an open, comprehensive, manually curated and interactive resource for studying experimentally observed post-translational modifications, primarily of human and mouse proteins. It encompasses 1,30,000 non-redundant modification sites, primarily phosphorylation, ubiquitinylation and acetylation. The interface is designed for clarity and ease of navigation. From the home page, users can launch simple or complex searches and browse high-throughput data sets by disease, tissue or cell line. Searches can be restricted by specific treatments, protein types, domains, cellular components, disease, cell types, cell lines, tissue and sequences or motifs. A few clicks of the mouse will take users to substrate pages or protein pages with sites, sequences, domain diagrams and molecular visualization of side-chains known to be modified; to site pages with information about how the modified site relates to the functions of specific proteins and cellular processes and to curated information pages summarizing the details from one record. PyMOL and Chimera scripts that colorize reactive groups on residues that are modified can be downloaded. Features designed to facilitate proteomic analyses include downloads of modification sites, kinase-substrate data sets, sequence logo generators, a Cytoscape plugin and BioPAX download to enable pathway visualization of the kinase-substrate interactions in PhosphoSitePlus®.
Figures




References
-
- Walsh CT, Garneau-Tsodikova S, Gatto GJ., Jr Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem. Int. Ed. Engl. 2005;44:7342–7372. - PubMed
-
- Mayer SE, Krebs EG. Studies on the phosphorylation and activation of skeletal muscle phosphorylase and phosphorylase kinase in vivo. J. Biol. Chem. 1970;245:3153–3160. - PubMed
-
- Varmus H, Hirai H, Morgan D, Kaplan J, Bishop JM. Function, location, and regulation of the src protein-tyrosine kinase. Princess Takamatsu Symp. 1989;20:63–70. - PubMed
-
- Sefton BM, Hunter T, Beemon K, Eckhart W. Evidence that the phosphorylation of tyrosine is essential for cellular transformation by Rous sarcoma virus. Cell. 1980;20:807–816. - PubMed
-
- Pearson RB, Kemp BE. Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol. 1991;200:62–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

