A novel zinc-finger nuclease platform with a sequence-specific cleavage module
- PMID: 22135304
- PMCID: PMC3315325
- DOI: 10.1093/nar/gkr1112
A novel zinc-finger nuclease platform with a sequence-specific cleavage module
Abstract
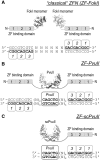
Zinc-finger nucleases (ZFNs) typically consist of three to four zinc fingers (ZFs) and the non-specific DNA-cleavage domain of the restriction endonuclease FokI. In this configuration, the ZFs constitute the binding module and the FokI domain the cleavage module. Whereas new binding modules, e.g. TALE sequences, have been considered as alternatives to ZFs, no efforts have been undertaken so far to replace the catalytic domain of FokI as the cleavage module in ZFNs. Here, we have fused a three ZF array to the restriction endonuclease PvuII to generate an alternative ZFN. While PvuII adds an extra element of specificity when combined with ZFs, ZF-PvuII constructs must be designed such that only PvuII sites with adjacent ZF-binding sites are cleaved. To achieve this, we introduced amino acid substitutions into PvuII that alter K(m) and k(cat) and increase fidelity. The optimized ZF-PvuII fusion constructs cleave DNA at addressed sites with a >1000-fold preference over unaddressed PvuII sites in vitro as well as in cellula. In contrast to the 'analogous' ZF-FokI nucleases, neither excess of enzyme over substrate nor prolonged incubation times induced unaddressed cleavage in vitro. These results present the ZF-PvuII platform as a valid alternative to conventional ZFNs.
Figures





Similar articles
-
Creating highly specific nucleases by fusion of active restriction endonucleases and catalytically inactive homing endonucleases.Nucleic Acids Res. 2012 Jan;40(2):847-60. doi: 10.1093/nar/gkr788. Epub 2011 Sep 29. Nucleic Acids Res. 2012. PMID: 21965534 Free PMC article.
-
TALE-PvuII fusion proteins--novel tools for gene targeting.PLoS One. 2013 Dec 5;8(12):e82539. doi: 10.1371/journal.pone.0082539. eCollection 2013. PLoS One. 2013. PMID: 24349308 Free PMC article.
-
Creating designed zinc-finger nucleases with minimal cytotoxicity.J Mol Biol. 2011 Jan 21;405(3):630-41. doi: 10.1016/j.jmb.2010.10.043. Epub 2010 Nov 19. J Mol Biol. 2011. PMID: 21094162 Free PMC article.
-
[Zinc finger nucleases and their application].Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2010 Apr;27(2):162-5. doi: 10.3760/cma.j.issn.1003-9406.2010.02.010. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2010. PMID: 20376797 Review. Chinese.
-
Zinc finger nucleases as tools to understand and treat human diseases.BMC Med. 2010 Jul 1;8:42. doi: 10.1186/1741-7015-8-42. BMC Med. 2010. PMID: 20594338 Free PMC article. Review.
Cited by
-
Programmable RNA-Guided RNA Effector Proteins Built from Human Parts.Cell. 2019 Jun 27;178(1):122-134.e12. doi: 10.1016/j.cell.2019.05.049. Epub 2019 Jun 20. Cell. 2019. PMID: 31230714 Free PMC article.
-
CRISPR-Cas Genome Surgery in Ophthalmology.Transl Vis Sci Technol. 2017 May 31;6(3):13. doi: 10.1167/tvst.6.3.13. eCollection 2017 May. Transl Vis Sci Technol. 2017. PMID: 28573077 Free PMC article. Review.
-
The I-TevI nuclease and linker domains contribute to the specificity of monomeric TALENs.G3 (Bethesda). 2014 Apr 16;4(6):1155-65. doi: 10.1534/g3.114.011445. G3 (Bethesda). 2014. PMID: 24739648 Free PMC article.
-
Compact designer TALENs for efficient genome engineering.Nat Commun. 2013;4:1762. doi: 10.1038/ncomms2782. Nat Commun. 2013. PMID: 23612303 Free PMC article.
-
Recent Advances in Genome-Engineering Strategies.Genes (Basel). 2023 Jan 2;14(1):129. doi: 10.3390/genes14010129. Genes (Basel). 2023. PMID: 36672870 Free PMC article. Review.
References
-
- Mackay JP, Segal DJ. Engineered Zinc Finger Proteins. New York: Humana Press; 2010.
-
- Wong GK, Chiu AT. Gene therapy, gene targeting and induced pluripotent stem cells: applications in monogenic disease treatment. Biotechnol. Adv. 2011;29:1–10. - PubMed
-
- Pingoud A, Silva GH. Precision genome surgery. Nat. Biotechnol. 2007;25:743–744. - PubMed
-
- Pingoud A, Wende W. Generation of novel nucleases with extended specificity by rational and combinatorial strategies. ChemBioChem. 2011;12:1495–1500. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

