Transcriptional regulatory circuitries in the human pathogen Candida albicans involving sense--antisense interactions
- PMID: 22135347
- PMCID: PMC3276616
- DOI: 10.1534/genetics.111.136267
Transcriptional regulatory circuitries in the human pathogen Candida albicans involving sense--antisense interactions
Abstract
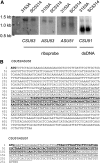
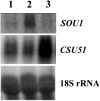
Candida albicans, a major human fungal pathogen, usually contains a diploid genome, but controls adaptation to a toxic alternative carbon source L-sorbose, by the reversible loss of one chromosome 5 (Ch5). We have previously identified multiple unique regions on Ch5 that repress the growth on sorbose. In one of the regions, the CSU51 gene determining the repressive property of the region was identified. We report here the identification of the CSU53 gene from a different region on Ch5. Most importantly, we find that CSU51 and CSU53 are associated with novel regulatory elements, ASUs, which are embedded within CSUs in an antisense configuration. ASUs act opposite to CSUs by enhancing the growth on sorbose. In respect to the CSU transcripts, the ASU long antisense transcripts are in lesser amounts, are completely overlapped, and are inversely related. ASUs interact with CSUs in natural CSU/ASU cis configurations, as well as when extra copies of ASUs are placed in trans to the CSU/ASU configurations. We suggest that ASU long embedded antisense transcripts modulate CSU sense transcripts.
Figures


References
-
- Ahmad A., Kabir M. A., Kravets A., Andaluz E., Larriba G., et al. , 2008. Chromosome instability and unusual features of some widely used strains of Candida albicans. Yeast 25: 433–448 - PubMed
-
- Camblong J., Iglesias N., Fickentscher C., Dieppois G., Stutz F., 2007. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in Saccharomyces cerevisiae. Cell 131: 706–717 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

