Genetic architecture of maize kernel composition in the nested association mapping and inbred association panels
- PMID: 22135431
- PMCID: PMC3271770
- DOI: 10.1104/pp.111.185033
Genetic architecture of maize kernel composition in the nested association mapping and inbred association panels
Abstract
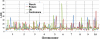
The maize (Zea mays) kernel plays a critical role in feeding humans and livestock around the world and in a wide array of industrial applications. An understanding of the regulation of kernel starch, protein, and oil is needed in order to manipulate composition to meet future needs. We conducted joint-linkage quantitative trait locus mapping and genome-wide association studies (GWAS) for kernel starch, protein, and oil in the maize nested association mapping population, composed of 25 recombinant inbred line families derived from diverse inbred lines. Joint-linkage mapping revealed that the genetic architecture of kernel composition traits is controlled by 21-26 quantitative trait loci. Numerous GWAS associations were detected, including several oil and starch associations in acyl-CoA:diacylglycerol acyltransferase1-2, a gene that regulates oil composition and quantity. Results from nested association mapping were verified in a 282 inbred association panel using both GWAS and candidate gene association approaches. We identified many beneficial alleles that will be useful for improving kernel starch, protein, and oil content.
Figures


References
-
- Ascenzi R, Gantt JS. (1997) A drought-stress-inducible histone gene in Arabidopsis thaliana is a member of a distinct class of plant linker histone variants. Plant Mol Biol 34: 629–641 - PubMed
-
- Barton NH, Keightley PD. (2002) Understanding quantitative genetic variation. Nat Rev Genet 3: 11–21 - PubMed
-
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B Met 57: 289–300
-
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 - PubMed
-
- Buchanan BB, Gruissem W, Jones RL. (2000) Biochemistry and Molecular Biology of Plants. American Society of Plant Biology, Rockville, MD
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

