RibM from Streptomyces davawensis is a riboflavin/roseoflavin transporter and may be useful for the optimization of riboflavin production strains
- PMID: 22136195
- PMCID: PMC3239331
- DOI: 10.1186/1472-6750-11-119
RibM from Streptomyces davawensis is a riboflavin/roseoflavin transporter and may be useful for the optimization of riboflavin production strains
Abstract
Background: The bacterium Bacillus subtilis, which is not a natural riboflavin overproducer, has been converted into an excellent production strain by classical mutagenesis and metabolic engineering. To our knowledge, the enhancement of riboflavin excretion from the cytoplasm of overproducing cells has not yet been considered as a target for (further) strain improvement. Here we evaluate the flavin transporter RibM from Streptomyces davawensis with respect to improvement of a riboflavin production strain.
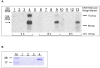
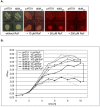
Results: The gene ribM from S. davawensis, coding for a putative facilitator of riboflavin uptake, was codon optimized (ribMopt) for expression in B. subtilis. The gene ribMopt was functionally introduced into B. subtilis using the isopropyl-β-thiogalactopyranoside (IPTG)-inducible expression plasmid pHT01: Northern-blot analysis of total RNA from IPTG treated recombinant B. subtilis cells revealed a ribMopt specific transcript. Western blot analysis showed that the his6-tagged heterologous gene product RibM was present in the cytoplasmic membrane. Expression of ribM in Escherichia coli increased [14C]riboflavin uptake, which was not affected by the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP). Expression of ribMopt supported growth of a B. subtilis ΔribB::Ermr ΔribU::Kanr double mutant deficient in riboflavin synthesis (ΔribB) and also deficient with respect to riboflavin uptake (ΔribU). Expression of ribMopt increased roseoflavin (a toxic riboflavin analog produced by S. davawensis) sensitivity of a B. subtilis ΔribU::Kanr strain. Riboflavin synthesis by a model riboflavin B. subtilis production strain overproducing RibM was increased significantly depending on the amount of the inducer IPTG.
Conclusions: The energy independent flavin facilitator RibM could in principle catalyze riboflavin export and thus may be useful to increase the riboflavin yield in a riboflavin production process using a recombinant RibM overproducing B. subtilis strain (or any other microorganism).
Figures



Similar articles
-
The RFN riboswitch of Bacillus subtilis is a target for the antibiotic roseoflavin produced by Streptomyces davawensis.RNA Biol. 2009 Jul-Aug;6(3):276-80. doi: 10.4161/rna.6.3.8342. Epub 2009 Jul 5. RNA Biol. 2009. PMID: 19333008
-
A second riboflavin import system is present in flavinogenic Streptomyces davaonensis and supports roseoflavin biosynthesis.Mol Microbiol. 2021 Aug;116(2):470-482. doi: 10.1111/mmi.14726. Epub 2021 May 6. Mol Microbiol. 2021. PMID: 33829573
-
The bifunctional flavokinase/flavin adenine dinucleotide synthetase from Streptomyces davawensis produces inactive flavin cofactors and is not involved in resistance to the antibiotic roseoflavin.J Bacteriol. 2008 Mar;190(5):1546-53. doi: 10.1128/JB.01586-07. Epub 2007 Dec 21. J Bacteriol. 2008. PMID: 18156273 Free PMC article.
-
Three biotechnical processes using Ashbya gossypii, Candida famata, or Bacillus subtilis compete with chemical riboflavin production.Appl Microbiol Biotechnol. 2000 May;53(5):509-16. doi: 10.1007/s002530051649. Appl Microbiol Biotechnol. 2000. PMID: 10855708 Review.
-
Biotechnology of riboflavin.Appl Microbiol Biotechnol. 2016 Mar;100(5):2107-19. doi: 10.1007/s00253-015-7256-z. Epub 2016 Jan 13. Appl Microbiol Biotechnol. 2016. PMID: 26758294 Review.
Cited by
-
Production of riboflavin and related cofactors by biotechnological processes.Microb Cell Fact. 2020 Feb 13;19(1):31. doi: 10.1186/s12934-020-01302-7. Microb Cell Fact. 2020. PMID: 32054466 Free PMC article. Review.
-
Identification and characterization of RibN, a novel family of riboflavin transporters from Rhizobium leguminosarum and other proteobacteria.J Bacteriol. 2013 Oct;195(20):4611-9. doi: 10.1128/JB.00644-13. Epub 2013 Aug 9. J Bacteriol. 2013. PMID: 23935051 Free PMC article.
-
Priming for welfare: gut microbiota is associated with equitation conditions and behavior in horse athletes.Sci Rep. 2020 May 20;10(1):8311. doi: 10.1038/s41598-020-65444-9. Sci Rep. 2020. PMID: 32433513 Free PMC article.
-
Underlying mechanisms for syntrophic metabolism of essential enzyme cofactors in microbial communities.ISME J. 2017 Jun;11(6):1434-1446. doi: 10.1038/ismej.2017.2. Epub 2017 Feb 10. ISME J. 2017. PMID: 28186498 Free PMC article.
-
Natural pigments derived from plants and microorganisms: classification, biosynthesis, and applications.Plant Biotechnol J. 2025 Feb;23(2):592-614. doi: 10.1111/pbi.14522. Epub 2024 Dec 6. Plant Biotechnol J. 2025. PMID: 39642082 Free PMC article. Review.
References
-
- Burgess CM, Slotboom DJ, Geertsma ER, Duurkens RH, Poolman B, van Sinderen D. The riboflavin transporter RibU in Lactococcus lactis: molecular characterization of gene expression and the transport mechanism. J Bacteriol. 2006;188(8):2752–2760. doi: 10.1128/JB.188.8.2752-2760.2006. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

