Structural basis for promoter-10 element recognition by the bacterial RNA polymerase σ subunit
- PMID: 22136875
- PMCID: PMC3245737
- DOI: 10.1016/j.cell.2011.10.041
Structural basis for promoter-10 element recognition by the bacterial RNA polymerase σ subunit
Abstract
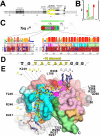
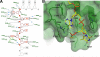
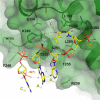
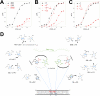
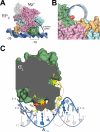
The key step in bacterial promoter opening is recognition of the -10 promoter element (T(-12)A(-11)T(-10)A(-9)A(-8)T(-7) consensus sequence) by the RNA polymerase σ subunit. We determined crystal structures of σ domain 2 bound to single-stranded DNA bearing-10 element sequences. Extensive interactions occur between the protein and the DNA backbone of every -10 element nucleotide. Base-specific interactions occur primarily with A(-11) and T(-7), which are flipped out of the single-stranded DNA base stack and buried deep in protein pockets. The structures, along with biochemical data, support a model where the recognition of the -10 element sequence drives initial promoter opening as the bases of the nontemplate strand are extruded from the DNA double-helix and captured by σ. These results provide a detailed structural basis for the critical roles of A(-11) and T(-7) in promoter melting and reveal important insights into the initiation of transcription bubble formation.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

Comment in
-
Lock and key to transcription: σ-DNA interaction.Cell. 2011 Dec 9;147(6):1218-9. doi: 10.1016/j.cell.2011.11.033. Cell. 2011. PMID: 22153066
References
-
- Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA. Structure of the bacterial RNA polymerase promoter specificity sigma factor. Mol Cell. 2002;9:527–539. - PubMed
-
- Chen YF, Helmann JD. The Bacillus subtilis flagellar regulatory protein σD: Overproduction, domain analysis and DNA-binding properties. J Mol Biol. 1995;249:743–753. - PubMed
-
- Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn 5 synaptic complex transposition intermediate. Science. 2000;289:77–85. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

