The H2A-H2B dimeric kinetic intermediate is stabilized by widespread hydrophobic burial with few fully native interactions
- PMID: 22137897
- PMCID: PMC3259194
- DOI: 10.1016/j.jmb.2011.11.032
The H2A-H2B dimeric kinetic intermediate is stabilized by widespread hydrophobic burial with few fully native interactions
Abstract
The H2A-H2B histone heterodimer folds via monomeric and dimeric kinetic intermediates. Within ∼5 ms, the H2A and H2B polypeptides associate in a nearly diffusion limited reaction to form a dimeric ensemble, denoted I₂ and I₂*, the latter being a subpopulation characterized by a higher content of nonnative structure (NNS). The I₂ ensemble folds to the native heterodimer, N₂, through an observable, first-order kinetic phase. To determine the regions of structure in the I₂ ensemble, we characterized 26 Ala mutants of buried hydrophobic residues, spanning the three helices of the canonical histone folds of H2A and H2B and the H2B C-terminal helix. All but one targeted residue contributed significantly to the stability of I₂, the transition state and N₂; however, only residues in the hydrophobic core of the dimer interface perturbed the I₂* population. Destabilization of I₂* correlated with slower folding rates, implying that NNS is not a kinetic trap but rather accelerates folding. The pattern of Φ values indicated that residues forming intramolecular interactions in the peripheral helices contributed similar stability to I₂ and N₂, but residues involved in intermolecular interactions in the hydrophobic core are only partially folded in I₂. These findings suggest a dimerize-then-rearrange model. Residues throughout the histone fold contribute to the stability of I₂, but after the rapid dimerization reaction, the hydrophobic core of the dimer interface has few fully native interactions. In the transition state leading to N₂, more native-like interactions are developed and nonnative interactions are rearranged.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures







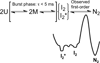
Similar articles
-
Mutational studies uncover non-native structure in the dimeric kinetic intermediate of the H2A-H2B heterodimer.J Mol Biol. 2010 Aug 20;401(3):518-31. doi: 10.1016/j.jmb.2010.06.034. Epub 2010 Jun 23. J Mol Biol. 2010. PMID: 20600120 Free PMC article.
-
Three-state kinetic folding mechanism of the H2A/H2B histone heterodimer: the N-terminal tails affect the transition state between a dimeric intermediate and the native dimer.J Mol Biol. 2005 Jan 28;345(4):827-36. doi: 10.1016/j.jmb.2004.11.006. J Mol Biol. 2005. PMID: 15588829
-
Mutational analysis of the stability of the H2A and H2B histone monomers.J Mol Biol. 2008 Dec 31;384(5):1369-83. doi: 10.1016/j.jmb.2008.10.040. Epub 2008 Oct 21. J Mol Biol. 2008. PMID: 18976667 Free PMC article.
-
Nucleosome adaptability conferred by sequence and structural variations in histone H2A-H2B dimers.Curr Opin Struct Biol. 2015 Jun;32:48-57. doi: 10.1016/j.sbi.2015.02.004. Epub 2015 Feb 27. Curr Opin Struct Biol. 2015. PMID: 25731851 Free PMC article. Review.
-
Archaeal histones: structures, stability and DNA binding.Biochem Soc Trans. 2004 Apr;32(Pt 2):227-30. doi: 10.1042/bst0320227. Biochem Soc Trans. 2004. PMID: 15046577 Review.
References
-
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. - PubMed
-
- Luger K. Dynamic nucleosomes. Chromosome Res. 2006;14:5–16. - PubMed
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
-
- Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–275. - PubMed
-
- Smith CL, Peterson CL. ATP-dependent chromatin remodeling. Curr Top Dev Biol. 2005;65:115–148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

