Repair of persistent strand breaks in the mitochondrial genome
- PMID: 22138376
- PMCID: PMC4586262
- DOI: 10.1016/j.mad.2011.11.003
Repair of persistent strand breaks in the mitochondrial genome
Abstract
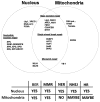
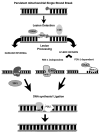
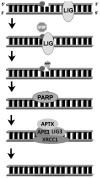
Oxidative DNA damage has been attributed to increased cancer incidence and premature aging phenotypes. Reactive oxygen species (ROS) are unavoidable byproducts of oxidative phosphorylation and are the major contributors of endogenous oxidative damage. To prevent the negative effects of ROS, cells have developed DNA repair mechanisms designed to specifically combat endogenous DNA modifications. The base excision repair (BER) pathway is primarily responsible for the repair of small non-helix distorting lesions and DNA single strand breaks. This repair pathway is found in all organisms, and in mammalian cells, consists of three related sub-pathways: short patch (SP-BER), long patch (LP-BER) and single strand break repair (SSBR). While much is known about nuclear BER, comparatively little is known about this pathway in the mitochondria, particularly the LP-BER and SSBR sub-pathways. There are a number of proteins that have recently been found to be involved in mitochondrial BER, including Cockayne syndrome proteins A and B (CSA and CSB), aprataxin (APTX), tryosyl-DNA phosphodiesterase 1 (TDP1), flap endonuclease 1 (FEN-1) and exonuclease G (EXOG). These significant advances in mitochondrial DNA repair may open new avenues in the management and treatment of a number of neurological disorders associated with mitochondrial dysfunction, and will be reviewed in further detail herein.
Copyright © 2011. Published by Elsevier Ireland Ltd.
Figures
References
-
- Aamann MD, Sorensen MM, Hvitby C, Berquist BR, Muftuoglu M, Tian J, de Souza-Pinto NC, Scheibye-Knudsen M, Wilson DM, III, Stevnsner T, Bohr VA. Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J. 2010;24:2334–2346. - PMC - PubMed
-
- Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, Caldecott KW, West SC. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. - PubMed
-
- Akbari M, Visnes T, Krokan HE, Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst) 2008;7:605–616. - PubMed
-
- Bentley D, Selfridge J, Millar JK, Samuel K, Hole N, Ansell JD, Melton DW. DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability. Nat Genet. 1996;13:489–491. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

