Lactoferricin B inhibits the phosphorylation of the two-component system response regulators BasR and CreB
- PMID: 22138548
- PMCID: PMC3322574
- DOI: 10.1074/mcp.M111.014720
Lactoferricin B inhibits the phosphorylation of the two-component system response regulators BasR and CreB
Abstract
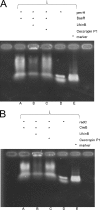
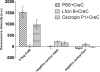
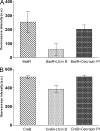
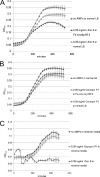
Natural antimicrobial peptides provide fundamental protection for multicellular organisms from microbes, such as Lactoferricin B (Lfcin B). Many studies have shown that Lfcin B penetrates the cell membrane and has intracellular activities. To elucidate the intracellular behavior of Lfcin B, we first used Escherichia coli K12 proteome chips to identify the intracellular targets of Lfcin B. The results showed that Lfcin B binds to two response regulators, BasR and CreB, of the two-component system. For further analysis, we conducted several in vitro and in vivo experiments and utilized bioinformatics methods. The electrophoretic mobility shift assays and kinase assays indicate that Lfcin B inhibits the phosphorylation of the response regulators (BasR and CreB) and their cognate sensor kinases (BasS and CreC). Antibacterial assays showed that Lfcin B reduced E. coli's tolerance to environmental stimuli, such as excessive ferric ions and minimal medium conditions. This is the first study to show that an antimicrobial peptide inhibits the growth of bacteria by influencing the phosphorylation of a two-component system directly.
Figures


Similar articles
-
Systematical Analysis of the Protein Targets of Lactoferricin B and Histatin-5 Using Yeast Proteome Microarrays.Int J Mol Sci. 2019 Aug 28;20(17):4218. doi: 10.3390/ijms20174218. Int J Mol Sci. 2019. PMID: 31466342 Free PMC article.
-
Identification of lactoferricin B intracellular targets using an Escherichia coli proteome chip.PLoS One. 2011;6(12):e28197. doi: 10.1371/journal.pone.0028197. Epub 2011 Dec 2. PLoS One. 2011. PMID: 22164243 Free PMC article.
-
Lactoferricin B causes depolarization of the cytoplasmic membrane of Escherichia coli ATCC 25922 and fusion of negatively charged liposomes.FEBS Lett. 2001 Mar 9;492(1-2):62-5. doi: 10.1016/s0014-5793(01)02233-5. FEBS Lett. 2001. PMID: 11248238
-
Lactoferricin derived from milk protein lactoferrin.Curr Pharm Des. 2003;9(16):1277-87. doi: 10.2174/1381612033454829. Curr Pharm Des. 2003. PMID: 12769736 Review.
-
Lactoferrin: an iron-binding antimicrobial protein against Escherichia coli infection.Biometals. 2011 Aug;24(4):585-94. doi: 10.1007/s10534-011-9423-8. Epub 2011 Feb 16. Biometals. 2011. PMID: 21327478 Review.
Cited by
-
Testing Antimicrobial Properties of Human Lactoferrin-Derived Fragments.Int J Mol Sci. 2023 Jun 23;24(13):10529. doi: 10.3390/ijms241310529. Int J Mol Sci. 2023. PMID: 37445717 Free PMC article.
-
Intracellular Targeting Mechanisms by Antimicrobial Peptides.Antimicrob Agents Chemother. 2017 Mar 24;61(4):e02340-16. doi: 10.1128/AAC.02340-16. Print 2017 Apr. Antimicrob Agents Chemother. 2017. PMID: 28167546 Free PMC article. Review.
-
Milk-Derived Antimicrobial Peptides: Overview, Applications, and Future Perspectives.Probiotics Antimicrob Proteins. 2023 Feb;15(1):44-62. doi: 10.1007/s12602-022-10004-y. Epub 2022 Nov 11. Probiotics Antimicrob Proteins. 2023. PMID: 36357656 Free PMC article. Review.
-
Characteristics and therapeutic applications of antimicrobial peptides.Biophys Rev (Melville). 2021 Feb 19;2(1):011301. doi: 10.1063/5.0035731. eCollection 2021 Mar. Biophys Rev (Melville). 2021. PMID: 38505398 Free PMC article. Review.
-
A High-Throughput Method for Identifying Novel Genes That Influence Metabolic Pathways Reveals New Iron and Heme Regulation in Pseudomonas aeruginosa.mSystems. 2021 Feb 2;6(1):e00933-20. doi: 10.1128/mSystems.00933-20. mSystems. 2021. PMID: 33531406 Free PMC article.
References
-
- Zasloff M. (2002) Antimicrobial peptides of multicellular organisms. Nature 415, 389–395 - PubMed
-
- Yamamoto K., Hirao K., Oshima T., Aiba H., Utsumi R., Ishihama A. (2005) Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 280, 1448–1456 - PubMed
-
- Hagiwara D., Yamashino T., Mizuno T. (2004) A Genome-wide view of the Escherichia coli BasS-BasR two-component system implicated in iron-responses. Biosci Biotechnol Biochem. 68, 1758–1767 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

