Genome-scale metabolic reconstruction and hypothesis testing in the methanogenic archaeon Methanosarcina acetivorans C2A
- PMID: 22139506
- PMCID: PMC3272958
- DOI: 10.1128/JB.06040-11
Genome-scale metabolic reconstruction and hypothesis testing in the methanogenic archaeon Methanosarcina acetivorans C2A
Abstract
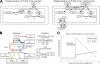
Methanosarcina acetivorans strain C2A is a marine methanogenic archaeon notable for its substrate utilization, genetic tractability, and novel energy conservation mechanisms. To help probe the phenotypic implications of this organism's unique metabolism, we have constructed and manually curated a genome-scale metabolic model of M. acetivorans, iMB745, which accounts for 745 of the 4,540 predicted protein-coding genes (16%) in the M. acetivorans genome. The reconstruction effort has identified key knowledge gaps and differences in peripheral and central metabolism between methanogenic species. Using flux balance analysis, the model quantitatively predicts wild-type phenotypes and is 96% accurate in knockout lethality predictions compared to currently available experimental data. The model was used to probe the mechanisms and energetics of by-product formation and growth on carbon monoxide, as well as the nature of the reaction catalyzed by the soluble heterodisulfide reductase HdrABC in M. acetivorans. The genome-scale model provides quantitative and qualitative hypotheses that can be used to help iteratively guide additional experiments to further the state of knowledge about methanogenesis.
Figures

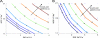

References
-
- Alberty RA. 2003. Thermodynamics of biochemical reactions. Massachusetts Institute of Technology, Cambridge, MA
-
- Anderson B, et al. April 2010. Methane and nitrous oxide emissions from natural sources. EPA 430-R-10-001. Office of Atmospheric Programs, Environmental Protection Agency, Washington, DC
-
- Aronson PS. 1985. Kinetic properties of the plasma membrane Na+-H+ exchanger. Annu. Rev. Physiol. 47:545–560 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

