neXtProt: a knowledge platform for human proteins
- PMID: 22139911
- PMCID: PMC3245017
- DOI: 10.1093/nar/gkr1179
neXtProt: a knowledge platform for human proteins
Abstract
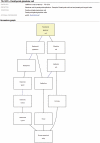
neXtProt (http://www.nextprot.org/) is a new human protein-centric knowledge platform. Developed at the Swiss Institute of Bioinformatics (SIB), it aims to help researchers answer questions relevant to human proteins. To achieve this goal, neXtProt is built on a corpus containing both curated knowledge originating from the UniProtKB/Swiss-Prot knowledgebase and carefully selected and filtered high-throughput data pertinent to human proteins. This article presents an overview of the database and the data integration process. We also lay out the key future directions of neXtProt that we consider the necessary steps to make neXtProt the one-stop-shop for all research projects focusing on human proteins.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

