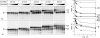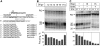Structural transitions in the transcription elongation complexes of bacterial RNA polymerase during σ-dependent pausing
- PMID: 22140106
- PMCID: PMC3326312
- DOI: 10.1093/nar/gkr1158
Structural transitions in the transcription elongation complexes of bacterial RNA polymerase during σ-dependent pausing
Abstract
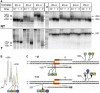
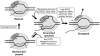
A transcription initiation factor, the σ(70) subunit of Escherichia coli RNA polymerase (RNAP) induces transcription pausing through the binding to a promoter-like pause-inducing sequence in the DNA template during transcription elongation. Here, we investigated the mechanism of σ-dependent pausing using reconstituted transcription elongation complexes which allowed highly efficient and precisely controlled pause formation. We demonstrated that, following engagement of the σ subunit to the pause site, RNAP continues RNA synthesis leading to formation of stressed elongation complexes, in which the nascent RNA remains resistant to Gre-induced cleavage while the transcription bubble and RNAP footprint on the DNA template extend in downstream direction, likely accompanied by DNA scrunching. The stressed complexes can then either break σ-mediated contacts and continue elongation or isomerize to a backtracked conformation. Suppressing of the RNAP backtracking decreases pausing and increases productive elongation. On the contrary, core RNAP mutations that impair RNAP interactions with the downstream part of the DNA template stimulate pausing, presumably by destabilizing the stressed complexes. We propose that interplay between DNA scrunching and RNAP backtracking may have an essential role in transcription pausing and its regulation in various systems.
Figures