Transcriptome-wide discovery of circular RNAs in Archaea
- PMID: 22140119
- PMCID: PMC3326292
- DOI: 10.1093/nar/gkr1009
Transcriptome-wide discovery of circular RNAs in Archaea
Abstract
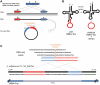
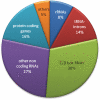
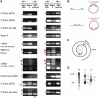
Circular RNA forms had been described in all domains of life. Such RNAs were shown to have diverse biological functions, including roles in the life cycle of viral and viroid genomes, and in maturation of permuted tRNA genes. Despite their potentially important biological roles, discovery of circular RNAs has so far been mostly serendipitous. We have developed circRNA-seq, a combined experimental/computational approach that enriches for circular RNAs and allows profiling their prevalence in a whole-genome, unbiased manner. Application of this approach to the archaeon Sulfolobus solfataricus P2 revealed multiple circular transcripts, a subset of which was further validated independently. The identified circular RNAs included expected forms, such as excised tRNA introns and rRNA processing intermediates, but were also enriched with non-coding RNAs, including C/D box RNAs and RNase P, as well as circular RNAs of unknown function. Many of the identified circles were conserved in Sulfolobus acidocaldarius, further supporting their functional significance. Our results suggest that circular RNAs, and particularly circular non-coding RNAs, are more prevalent in archaea than previously recognized, and might have yet unidentified biological roles. Our study establishes a specific and sensitive approach for identification of circular RNAs using RNA-seq, and can readily be applied to other organisms.
Figures

References
-
- Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558–560. - PubMed
-
- Flores R, Hernandez C, Martinez de Alba AE, Daros JA, Di Serio F. Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 2005;43:117–139. - PubMed
-
- Soma A, Onodera A, Sugahara J, Kanai A, Yachie N, Tomita M, Kawamura F, Sekine Y. Permuted tRNA genes expressed via a circular RNA intermediate in Cyanidioschyzon merolae. Science. 2007;318:450–453. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

