Snf1-like protein kinase Ssp2 regulates glucose derepression in Schizosaccharomyces pombe
- PMID: 22140232
- PMCID: PMC3272901
- DOI: 10.1128/EC.05268-11
Snf1-like protein kinase Ssp2 regulates glucose derepression in Schizosaccharomyces pombe
Abstract
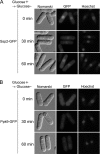
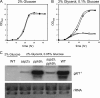
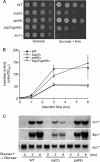
The function of two fission yeast genes, SPCC74.03c/ssp2(+) and SPAC23H4.02/ppk9(+), encoding an Snf1-like protein kinase were investigated. Deletion of ssp2(+) caused a partial defect in glucose derepression of inv1(+), fbp1(+), and gld1(+) and in assimilation of sucrose and glycerol, while a mutation in ppk9(+) had no apparent effect. Scr1, a transcription factor involved in glucose repression, localized to the nucleus under glucose-rich conditions and to the cytoplasm during glucose starvation in wild-type cells. In contrast, in the ssp2Δ mutant, Scr1 localized to the nucleus in cells grown in glucose-rich medium as well as in glucose-starved cells. Immunoblot analysis showed that Ssp2 is required for the phosphorylation of Scr1 upon glucose deprivation. Mutation of five putative Ssp2 recognition sites in Scr1 prevented glucose derepression of invertase in glucose-starved cells. These results indicate that Ssp2 regulates phosphorylation and subcellular localization of Scr1 in response to glucose.
Figures



References
-
- Carlson M. 1999. Glucose repression in yeast. Curr. Opin. Microbiol. 2:202–207 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

