Copy number variation of KIR genes influences HIV-1 control
- PMID: 22140359
- PMCID: PMC3226550
- DOI: 10.1371/journal.pbio.1001208
Copy number variation of KIR genes influences HIV-1 control
Erratum in
- PLoS Biol. 2011 Dec;9(12). doi:10.1371/annotation/7e17b146-a69c-4e83-9230-7340486d9dc8
Abstract
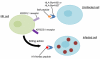
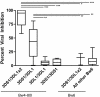
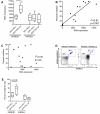
A genome-wide screen for large structural variants showed that a copy number variant (CNV) in the region encoding killer cell immunoglobulin-like receptors (KIR) associates with HIV-1 control as measured by plasma viral load at set point in individuals of European ancestry. This CNV encompasses the KIR3DL1-KIR3DS1 locus, encoding receptors that interact with specific HLA-Bw4 molecules to regulate the activation of lymphocyte subsets including natural killer (NK) cells. We quantified the number of copies of KIR3DS1 and KIR3DL1 in a large HIV-1 positive cohort, and showed that an increase in KIR3DS1 count associates with a lower viral set point if its putative ligand is present (p = 0.00028), as does an increase in KIR3DL1 count in the presence of KIR3DS1 and appropriate ligands for both receptors (p = 0.0015). We further provide functional data that demonstrate that NK cells from individuals with multiple copies of KIR3DL1, in the presence of KIR3DS1 and the appropriate ligands, inhibit HIV-1 replication more robustly, and associated with a significant expansion in the frequency of KIR3DS1+, but not KIR3DL1+, NK cells in their peripheral blood. Our results suggest that the relative amounts of these activating and inhibitory KIR play a role in regulating the peripheral expansion of highly antiviral KIR3DS1+ NK cells, which may determine differences in HIV-1 control following infection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Bashirova A. A, Martin M. P, McVicar D. W, Carrington M. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu Rev Genomics Hum Genet. 2006;7:277–300. - PubMed
-
- Uhrberg M, Valiante N. M, Shum B. P, Shilling H. G, Lienert-Weidenbach K, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. - PubMed
-
- Martin M. P, Bashirova A, Traherne J, Trowsdale J, Carrington M. Cutting Edge: expansion of the KIR locus by unequal crossing over. J Immunol. 2003;171:2192–1295. - PubMed
-
- Williams F, Maxwell L. D, Halfpenny I. A, Meenagh A, Sleator C, et al. Multiple copies of KIR 3DL/S1 and KIR 2DL4 genes identified in a number of individuals. Hum Immunol. 2003;64:729–732. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- U01 AI035042/AI/NIAID NIH HHS/United States
- U01AIO67854/PHS HHS/United States
- U01-AI-37613/AI/NIAID NIH HHS/United States
- 5-M01-RR-00052/RR/NCRR NIH HHS/United States
- 5 T32 GM007754-29/GM/NIGMS NIH HHS/United States
- U01 AI035041/AI/NIAID NIH HHS/United States
- M01 RR000052/RR/NCRR NIH HHS/United States
- U01-AI-37984/AI/NIAID NIH HHS/United States
- U01 AI035043/AI/NIAID NIH HHS/United States
- U01-AI-35039/AI/NIAID NIH HHS/United States
- U01 AI035040/AI/NIAID NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- U01 AI037984/AI/NIAID NIH HHS/United States
- U01-AI-35043/AI/NIAID NIH HHS/United States
- U01-AI-35042/AI/NIAID NIH HHS/United States
- T32 GM007754/GM/NIGMS NIH HHS/United States
- U01 AI037613/AI/NIAID NIH HHS/United States
- U01-AI-35041/AI/NIAID NIH HHS/United States
- U01-AI-35040/AI/NIAID NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- U01 AI035039/AI/NIAID NIH HHS/United States
- P30 AI060354/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

