A systematic analysis of the 3'UTR of HNF4A mRNA reveals an interplay of regulatory elements including miRNA target sites
- PMID: 22140441
- PMCID: PMC3227676
- DOI: 10.1371/journal.pone.0027438
A systematic analysis of the 3'UTR of HNF4A mRNA reveals an interplay of regulatory elements including miRNA target sites
Abstract
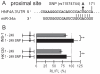
Dysfunction of hepatocyte nuclear factor 4α (HNF4α) has been linked to maturity onset diabetes of the young (MODY1), diabetes type II and possibly to renal cell carcinoma (RCC). Whereas diabetes causing mutations are well known, there are no HNF4A mutations found in RCC. Since so far analyses have been constricted to the promoter and open reading frame of HNF4A, we performed a systematic analysis of the human HNF4A 3'UTR. We identified a short (1724 nt) and long (3180 nt) 3'UTR that are much longer than the open reading frame and conferred a repressive effect in luciferase reporter assays in HEK293 and INS-1 cells. By dissecting the 3'UTR into several pieces, we located two distinct elements of about 400 nt conferring a highly repressive effect. These negative elements A and B are counteracted by a balancer element of 39 nt located within the 5' end of the HNF4A 3'UTR. Dicer knock-down experiments implied that the HNF4A 3'UTR is regulated by miRNAs. More detailed analysis showed that miR-34a and miR-21 both overexpressed in RCC cooperate in downregulation of the HNF4A mRNA. One of the identified miR-34a binding sites is destroyed by SNP rs11574744. The identification of several regulatory elements within the HNF4A 3'UTR justifies the analysis of the 3'UTR sequence to explore the dysfunction of HNF4α in diabetes and RCC.
Conflict of interest statement
Figures

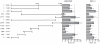


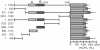

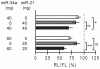

Similar articles
-
In silico and in vitro identification of microRNAs that regulate hepatic nuclear factor 4α expression.Drug Metab Dispos. 2012 Apr;40(4):726-33. doi: 10.1124/dmd.111.040329. Epub 2012 Jan 9. Drug Metab Dispos. 2012. PMID: 22232426 Free PMC article.
-
An 11-nt sequence polymorphism at the 3'UTR of human SFTPA1 and SFTPA2 gene variants differentially affect gene expression levels and miRNA regulation in cell culture.Am J Physiol Lung Cell Mol Physiol. 2014 Jul 1;307(1):L106-19. doi: 10.1152/ajplung.00313.2013. Epub 2014 May 2. Am J Physiol Lung Cell Mol Physiol. 2014. PMID: 24793167 Free PMC article.
-
The role of the 3'UTR region in the regulation of the ACVR1/Alk-2 gene expression.PLoS One. 2012;7(12):e50958. doi: 10.1371/journal.pone.0050958. Epub 2012 Dec 5. PLoS One. 2012. PMID: 23227223 Free PMC article.
-
3'UTR Diversity: Expanding Repertoire of RNA Alterations in Human mRNAs.Mol Cells. 2023 Jan 31;46(1):48-56. doi: 10.14348/molcells.2023.0003. Epub 2023 Jan 20. Mol Cells. 2023. PMID: 36697237 Free PMC article. Review.
-
The emerging theme of 3'UTR mRNA isoform regulation in reprogramming of cell metabolism.Biochem Soc Trans. 2023 Jun 28;51(3):1111-1119. doi: 10.1042/BST20221128. Biochem Soc Trans. 2023. PMID: 37171086 Free PMC article. Review.
Cited by
-
MicroRNA-34a modulates MDM4 expression via a target site in the open reading frame.PLoS One. 2012;7(8):e42034. doi: 10.1371/journal.pone.0042034. Epub 2012 Aug 1. PLoS One. 2012. PMID: 22870278 Free PMC article.
-
Evaluation of miRNA-binding-site SNPs of MRE11A, NBS1, RAD51 and RAD52 involved in HRR pathway genes and risk of breast cancer in China.Mol Genet Genomics. 2015 Jun;290(3):1141-53. doi: 10.1007/s00438-014-0983-5. Epub 2015 Jan 9. Mol Genet Genomics. 2015. PMID: 25566853
-
Noncoding microRNAs: small RNAs play a big role in regulation of ADME?Acta Pharm Sin B. 2012 Apr;2(2):93-101. doi: 10.1016/j.apsb.2012.02.011. Epub 2012 Mar 31. Acta Pharm Sin B. 2012. PMID: 32154096 Free PMC article.
-
The network of microRNAs, transcription factors, target genes and host genes in human renal cell carcinoma.Oncol Lett. 2015 Jan;9(1):498-506. doi: 10.3892/ol.2014.2683. Epub 2014 Nov 7. Oncol Lett. 2015. PMID: 25436016 Free PMC article.
-
The role of microRNAs in hepatocyte nuclear factor-4alpha expression and transactivation.Biochim Biophys Acta. 2013 May;1829(5):436-42. doi: 10.1016/j.bbagrm.2012.12.009. Epub 2013 Jan 5. Biochim Biophys Acta. 2013. PMID: 23298640 Free PMC article.
References
-
- Bolotin E, Schnabl JM, Sladek FM. HNF4A (Homo sapiens). Transcription Factor Encyclopedia. 2010. Available: http://www.cisreg.ca/tfe. Accessed 2010 July 15.
-
- Bolotin E, Sladek FM, Schnabl JM. Hnf4a (Mus musculus). Transcription Factor Encyclopedia. 2010. Available: http://www.cisreg.ca/tfe. Accessed 2010 July 15.
-
- Erdmann S, Senkel S, Arndt T, Lucas B, Lausen J, et al. Tissue-specific transcription factor HNF4alpha inhibits cell proliferation and induces apoptosis in the pancreatic INS-1 beta-cell line. Biol Chem. 2007;388:91–106. - PubMed
-
- Grigo K, Wirsing A, Lucas B, Klein-Hitpass L, Ryffel GU. HNF4alpha orchestrates a set of 14 genes to down-regulate cell proliferation in kidney cells. Biol Chem. 2008;389:179–187. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

