Post-transcriptional regulation of PDGFα-receptor in O-2A progenitor cells
- PMID: 22140595
- PMCID: PMC3228579
Post-transcriptional regulation of PDGFα-receptor in O-2A progenitor cells
Abstract
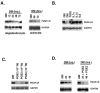
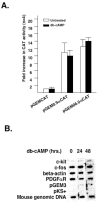
latelet-derived growth factor alpha-receptor (PDGFαR) mediated signaling plays a key role in the development of glial cells of the central nervous system. In vivo and in vitro studies show that PDGFαR is actively expressed in proliferative and motile oligodendrocyte type-2 astrocyte (O-2A) glial progenitor cells. However, PDGFαR expression is barely detectable in mature glial cells. The exact mechanism underlying the loss of PDGFαR expression is unknown. In this study, we employed a rat brain-derived O-2A glial progenitor cell line, CG4 as a culture model to investigate signals capable of inhibiting PDGFαR gene expression. PDGFαR mRNA levels decreased significantly as CG4 cells differentiated into both oligodendrocyte and astrocyte lineages. We showed that inhibition of PDGFαR expression was promoted by prostaglandin E2 via protein kinase A activation. Both cAMP analogs (db-cAMP and 8'bromo-cAMP) and adenylate cyclase activator (forskolin) were potent suppressors of PDGFαR expression in CG4 cells. This inhibitory effect resulted from an increased destabilization of PDGFαR mRNA instead of a decreased PDGFαR gene transcription. Importantly, db-cAMP failed to reduce PDGFαR mRNA levels in several PDGFαR over-expressing human glioma cell lines. Together, these results suggest that cAMP-dependent pathway played a key regulatory role in controlling PDGFαR mRNA levels during normal glial development, and that a breakdown in the cross talk between cAMP and PDGF pathways may underlie the uncontrolled proliferation and immature differentiation state in the glial tumors.
Keywords: PDGF; cyclic AMP; glioma; mRNA turnover.
Figures



Similar articles
-
Transcriptional regulation of the platelet-derived growth factor alpha receptor gene via CCAAT/enhancer-binding protein-delta in vascular smooth muscle cells.J Biol Chem. 1999 Sep 3;274(36):25576-82. doi: 10.1074/jbc.274.36.25576. J Biol Chem. 1999. PMID: 10464291
-
Targeted overexpression of CCAAT/enhancer-binding protein-delta evokes enhanced gene transcription of platelet-derived growth factor alpha-receptor in vascular smooth muscle cells.Circ Res. 2001 Sep 14;89(6):503-8. doi: 10.1161/hh1801.096265. Circ Res. 2001. PMID: 11557737
-
Autocrine inhibition of mitotic activity in cultured oligodendrocyte-type-2 astrocyte (O-2A) precursor cells.Glia. 1992;6(1):30-8. doi: 10.1002/glia.440060105. Glia. 1992. PMID: 1387386
-
Up-regulation of the cAMP/PKA pathway inhibits proliferation, induces differentiation, and leads to apoptosis in malignant gliomas.Lab Invest. 1998 Feb;78(2):165-74. Lab Invest. 1998. PMID: 9484714 Review.
-
From rodent glial precursor cell to human glial neoplasia in the oligodendrocyte-type-2 astrocyte lineage.Glia. 1995 Nov;15(3):222-30. doi: 10.1002/glia.440150304. Glia. 1995. PMID: 8586459 Review.
References
-
- Bergsten E, Uutela M, Li X, Pietras K, Ostman A, Heldin CH, Alitalo K, Eriksson U. PDGF-D is a specific, protease-activated ligand for the PDGF beta-receptor. Nat Cell Biol. 2001;3:512–516. - PubMed
-
- Bowen Pope DF, Hart CE, Seifert RA. Sera and conditioned media contain different isoforms of platelet-derived growth factor (PDGF) which bind to different class of PDGF receptor. J Biol Chem. 1983;254:2502–2508. - PubMed
-
- Doolittle RF, Hunkapiller MW, Hood LE, Devare SG, Robbins KC, Aaronson SA, Antoniades HN. Simian sarcoma virus oncogene, v-sis, is derived from the gene (or genes) encoding a platelet-derived growth factor. Science. 1983;221:275–277. - PubMed
-
- LaRochelle WJ, Jeffers M, McDonald WF, Chillakuru RA, Giese NA, Lokker NA, Sullivan C, Boldog FL, Yang M, Vernet C, Burgess CE, Fernandes E, Deegler LL, Rittman B, Shimkets J, Shimkets RA, Rothberg JM, Lichenstein HS. PDGF-D, a new protease-activated growth factor. Nat Cell Biol. 2001;3:517–521. - PubMed
-
- Li X, Ponten A, Aase K, Karlsson L, Abramsson A, Uutela M, Backstrom G, Hellstrom M, Bostrom H, Li H, Soriano P, Betsholtz C, Heldin CH, Alitalo K, Ostman A, Eriksson U. PDGF-C is a new protease-activated ligand for the PDGF alpha-receptor. Nat Cell Biol. 2000;2:302–309. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
