Detection and quantification of poliovirus infection using FTIR spectroscopy and cell culture
- PMID: 22142483
- PMCID: PMC3260089
- DOI: 10.1186/1754-1611-5-16
Detection and quantification of poliovirus infection using FTIR spectroscopy and cell culture
Abstract
Background: In a globalized word, prevention of infectious diseases is a major challenge. Rapid detection of viable virus particles in water and other environmental samples is essential to public health risk assessment, homeland security and environmental protection. Current virus detection methods, especially assessing viral infectivity, are complex and time-consuming, making point-of-care detection a challenge. Faster, more sensitive, highly specific methods are needed to quantify potentially hazardous viral pathogens and to determine if suspected materials contain viable viral particles. Fourier transform infrared (FTIR) spectroscopy combined with cellular-based sensing, may offer a precise way to detect specific viruses. This approach utilizes infrared light to monitor changes in molecular components of cells by tracking changes in absorbance patterns produced following virus infection. In this work poliovirus (PV1) was used to evaluate the utility of FTIR spectroscopy with cell culture for rapid detection of infective virus particles.
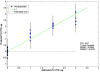
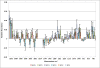
Results: Buffalo green monkey kidney (BGMK) cells infected with different virus titers were studied at 1 - 12 hours post-infection (h.p.i.). A partial least squares (PLS) regression method was used to analyze and model cellular responses to different infection titers and times post-infection. The model performs best at 8 h.p.i., resulting in an estimated root mean square error of cross validation (RMSECV) of 17 plaque forming units (PFU)/ml when using low titers of infection of 10 and 100 PFU/ml. Higher titers, from 103 to 106 PFU/ml, could also be reliably detected.
Conclusions: This approach to poliovirus detection and quantification using FTIR spectroscopy and cell culture could potentially be extended to compare biochemical cell responses to infection with different viruses. This virus detection method could feasibly be adapted to an automated scheme for use in areas such as water safety monitoring and medical diagnostics.
Figures
Similar articles
-
Detection of infective poliovirus by a simple, rapid, and sensitive flow cytometry method based on fluorescence resonance energy transfer technology.Appl Environ Microbiol. 2010 Jan;76(2):584-8. doi: 10.1128/AEM.01851-09. Epub 2009 Nov 20. Appl Environ Microbiol. 2010. PMID: 19933336 Free PMC article.
-
A virion concentration method for detection of human enteric viruses in oysters by PCR and oligoprobe hybridization.Appl Environ Microbiol. 1996 Jun;62(6):2074-80. doi: 10.1128/aem.62.6.2074-2080.1996. Appl Environ Microbiol. 1996. PMID: 8787405 Free PMC article.
-
Attenuated total reflectance-Fourier transform infrared (ATR-FTIR) spectroscopy coupled with chemometric analysis for detection and quantification of adulteration in lavender and citronella essential oils.Phytochem Anal. 2021 Nov;32(6):907-920. doi: 10.1002/pca.3034. Epub 2021 Feb 10. Phytochem Anal. 2021. PMID: 33565180
-
[Establishment of the Mathematical Model for PMI Estimation Using FTIR Spectroscopy and Data Mining Method].Fa Yi Xue Za Zhi. 2018 Feb;34(1):1-6. doi: 10.3969/j.issn.1004-5619.2018.01.001. Epub 2018 Feb 25. Fa Yi Xue Za Zhi. 2018. PMID: 29577696 Chinese.
-
Quantitative evaluation of multiple adulterants in roasted coffee by Diffuse Reflectance Infrared Fourier Transform Spectroscopy (DRIFTS) and chemometrics.Talanta. 2013 Oct 15;115:563-8. doi: 10.1016/j.talanta.2013.06.004. Epub 2013 Jun 14. Talanta. 2013. PMID: 24054633
Cited by
-
Enteroviruses in Water: Epidemiology, Detection and Inactivation.Environ Microbiol. 2025 May;27(5):e70109. doi: 10.1111/1462-2920.70109. Environ Microbiol. 2025. PMID: 40390239 Free PMC article. Review.
-
Potential of infrared microscopy to differentiate between dementia with Lewy bodies and Alzheimer's diseases using peripheral blood samples and machine learning algorithms.J Biomed Opt. 2020 Apr;25(4):1-15. doi: 10.1117/1.JBO.25.4.046501. J Biomed Opt. 2020. PMID: 32329265 Free PMC article.
-
Raman and fourier transform infrared spectroscopy techniques for detection of coronavirus (COVID-19): a mini review.Front Chem. 2023 May 18;11:1193030. doi: 10.3389/fchem.2023.1193030. eCollection 2023. Front Chem. 2023. PMID: 37273513 Free PMC article. Review.
-
ATR-FTIR spectroscopy and chemometrics as a quick and simple alternative for discrimination of SARS-CoV-2 infected food of animal origin.Spectrochim Acta A Mol Biomol Spectrosc. 2023 Jan 15;285:121883. doi: 10.1016/j.saa.2022.121883. Epub 2022 Sep 14. Spectrochim Acta A Mol Biomol Spectrosc. 2023. PMID: 36126622 Free PMC article.
-
Spectroscopy with computational analysis in virological studies: A decade (2006-2016).Trends Analyt Chem. 2017 Dec;97:244-256. doi: 10.1016/j.trac.2017.09.015. Epub 2017 Sep 21. Trends Analyt Chem. 2017. PMID: 32287542 Free PMC article. Review.
References
-
- Pruss-Ustun A, Bos R, Gore F, Bartram J. Safe water, better health: costs, benefits and sustainability of interventions to protect and promote health. World Health Organization, Geneva. 2008.
-
- Schiff GM, Stefanovic GM, Young B, Pennekamp JK. In: Enteric viruses in water. Melnick JL, editor. Vol. 15. 1984. Minimum human infectious dose of enteric viruses (echovirus-12) in drinking water; pp. 222–228.
-
- Non-polio enterovirus infections, Division of viral Diseases Centers for Disease Control and Prevention. http://www.cdc.gov/ncidod/dvrd/revb/enterovirus/non-polio_entero.htm last access Nov 2011.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

