Dynamic metabolic labeling of DNA in vivo with arabinosyl nucleosides
- PMID: 22143759
- PMCID: PMC3251089
- DOI: 10.1073/pnas.1101126108
Dynamic metabolic labeling of DNA in vivo with arabinosyl nucleosides
Abstract
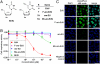
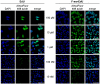
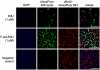
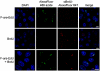
Commonly used metabolic labels for DNA, including 5-ethynyl-2'-deoxyuridine (EdU) and BrdU, are toxic antimetabolites that cause DNA instability, necrosis, and cell-cycle arrest. In addition to perturbing biological function, these properties can prevent metabolic labeling studies where subsequent tissue survival is needed. To bypass the metabolic pathways responsible for toxicity, while maintaining the ability to be metabolically incorporated into DNA, we synthesized and evaluated a small family of arabinofuranosyl-ethynyluracil derivatives. Among these, (2'S)-2'-deoxy-2'-fluoro-5-ethynyluridine (F-ara-EdU) exhibited selective DNA labeling, yet had a minimal impact on genome function in diverse tissue types. Metabolic incorporation of F-ara-EdU into DNA was readily detectable using copper(I)-catalyzed azide-alkyne "click" reactions with fluorescent azides. F-ara-EdU is less toxic than both BrdU and EdU, and it can be detected with greater sensitivity in experiments where long-term cell survival and/or deep-tissue imaging are desired. In contrast to previously reported 2'-arabino modified nucleosides and EdU, F-ara-EdU causes little or no cellular arrest or DNA synthesis inhibition. F-ara-EdU is therefore ideally suited for pulse-chase experiments aimed at "birth dating" DNA in vivo. As a demonstration, Zebrafish embryos were microinjected with F-ara-EdU at the one-cell stage and chased by BrdU at 10 h after fertilization. Following 3 d of development, complex patterns of quiescent/senescent cells containing only F-ara-EdU were observed in larvae along the dorsal side of the notochord and epithelia. Arabinosyl nucleoside derivatives therefore provide unique and effective means to introduce bioorthogonal functional groups into DNA for diverse applications in basic research, biotechnology, and drug discovery.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Comment in
-
Bearing the mark.Nat Methods. 2012 Feb;9(2):127. doi: 10.1038/nmeth.1885. Nat Methods. 2012. PMID: 22396970 No abstract available.
References
-
- Bucci M, Goodman C, Sheppard TL. A decade of chemical biology. Nat Chem Biol. 2010;6:847–854. - PubMed
-
- Zink D, Cremer T. Cell nucleus: Chromosome dynamics in nuclei of living cells. Curr Biol. 1998;8:R321–R324. - PubMed
-
- Farkan S. High-resolution autoradiography as a tool for the localization of nucleic acid synthesis and distribution in the mammalian cell nucleus. J Microsc. 1976;106:159–171. - PubMed
-
- Taupin P. BrdU immunohistochemistry for studying adult neurogenesis: Paradigms, pitfalls, limitations, and validation. Brain Res Rev. 2007;53:198–214. - PubMed
-
- Best MD. Click chemistry and bioorthogonal reactions: Unprecedented selectivity in the labeling of biological molecules. Biochemistry. 2009;48:6571–6584. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

