Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses
- PMID: 22143798
- PMCID: PMC3251064
- DOI: 10.1073/pnas.1113801108
Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses
Abstract
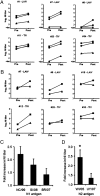
Seasonal epidemics caused by influenza virus are driven by antigenic changes (drift) in viral surface glycoproteins that allow evasion from preexisting humoral immunity. Antigenic drift is a feature of not only the hemagglutinin (HA), but also of neuraminidase (NA). We have evaluated the antigenic evolution of each protein in H1N1 and H3N2 viruses used in vaccine formulations during the last 15 y by analysis of HA and NA inhibition titers and antigenic cartography. As previously shown for HA, genetic changes in NA did not always lead to an antigenic change. The noncontinuous pattern of NA drift did not correspond closely with HA drift in either subtype. Although NA drift was demonstrated using ferret sera, we show that these changes also impact recognition by NA-inhibiting antibodies in human sera. Remarkably, a single point mutation in the NA of A/Brisbane/59/2007 was primarily responsible for the lack of inhibition by polyclonal antibodies specific for earlier strains. These data underscore the importance of NA inhibition testing to define antigenic drift when there are sequence changes in NA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



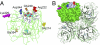
References
-
- Webster RG, Laver WG, Air GM, Schild GC. Molecular mechanisms of variation in influenza viruses. Nature. 1982;296:115–121. - PubMed
-
- Laver WG, Air GM, Webster RG, Markoff LJ. Amino acid sequence changes in antigenic variants of type A influenza virus N2 neuraminidase. Virology. 1982;122:450–460. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

