Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis
- PMID: 22143917
- PMCID: PMC3276081
- DOI: 10.1093/jxb/err315
Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis
Abstract
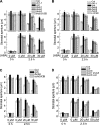
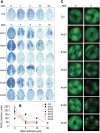
The light-harvesting chlorophyll a/b binding proteins (LHCB) are perhaps the most abundant membrane proteins in nature. It is reported here that the down-regulation or disruption of any member of the LHCB family, LHCB1, LHCB2, LHCB3, LHCB4, LHCB5, or LHCB6, reduces responsiveness of stomatal movement to ABA, and therefore results in a decrease in plant tolerance to drought stress in Arabidopsis thaliana. By contrast, over-expression of a LHCB member, LHCB6, enhances stomatal sensitivity to ABA. In addition, the reactive oxygen species (ROS) homeostasis and a set of ABA-responsive genes are altered in the lhcb mutants. These data demonstrate that LHCBs play a positive role in guard cell signalling in response to ABA and suggest that they may be involved in ABA signalling partly by modulating ROS homeostasis.
Figures



Similar articles
-
Light-harvesting chlorophyll a/b-binding proteins, positively involved in abscisic acid signalling, require a transcription repressor, WRKY40, to balance their function.J Exp Bot. 2013 Dec;64(18):5443-56. doi: 10.1093/jxb/ert307. Epub 2013 Sep 27. J Exp Bot. 2013. PMID: 24078667 Free PMC article.
-
Phospholipase dalpha1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis.Plant Cell. 2009 Aug;21(8):2357-77. doi: 10.1105/tpc.108.062992. Epub 2009 Aug 18. Plant Cell. 2009. PMID: 19690149 Free PMC article.
-
NADK2 positively modulates abscisic acid-induced stomatal closure by affecting accumulation of H2O2, Ca2+ and nitric oxide in Arabidopsis guard cells.Plant Sci. 2017 Sep;262:81-90. doi: 10.1016/j.plantsci.2017.06.003. Epub 2017 Jun 9. Plant Sci. 2017. PMID: 28716423
-
Overexpression of Arabidopsis acyl-CoA-binding protein ACBP2 enhances drought tolerance.Plant Cell Environ. 2013 Feb;36(2):300-14. doi: 10.1111/j.1365-3040.2012.02574.x. Epub 2012 Aug 8. Plant Cell Environ. 2013. PMID: 22788984
-
The power of small: microRNAs modulating stomatal movement.Trends Plant Sci. 2025 Feb;30(2):122-124. doi: 10.1016/j.tplants.2024.09.009. Epub 2024 Oct 1. Trends Plant Sci. 2025. PMID: 39358103 Review.
Cited by
-
Potential Role of Photosynthesis in the Regulation of Reactive Oxygen Species and Defence Responses to Blumeria graminis f. sp. tritici in Wheat.Int J Mol Sci. 2020 Aug 11;21(16):5767. doi: 10.3390/ijms21165767. Int J Mol Sci. 2020. PMID: 32796723 Free PMC article.
-
A Brassica napus Reductase Gene Dissected by Associative Transcriptomics Enhances Plant Adaption to Freezing Stress.Front Plant Sci. 2020 Jun 26;11:971. doi: 10.3389/fpls.2020.00971. eCollection 2020. Front Plant Sci. 2020. PMID: 32676095 Free PMC article.
-
Phylogenetic, Structural and Functional Evolution of the LHC Gene Family in Plant Species.Int J Mol Sci. 2022 Dec 28;24(1):488. doi: 10.3390/ijms24010488. Int J Mol Sci. 2022. PMID: 36613939 Free PMC article.
-
Guard cell photosynthesis is crucial in abscisic acid-induced stomatal closure.Plant Direct. 2019 May 30;3(5):e00137. doi: 10.1002/pld3.137. eCollection 2019 May. Plant Direct. 2019. PMID: 31245777 Free PMC article.
-
The beta Subunit of Nascent Polypeptide Associated Complex Plays A Role in Flowers and Siliques Development of Arabidopsis thaliana.Int J Mol Sci. 2020 Mar 17;21(6):2065. doi: 10.3390/ijms21062065. Int J Mol Sci. 2020. PMID: 32192231 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

