Conserved molecular interactions within the HBO1 acetyltransferase complexes regulate cell proliferation
- PMID: 22144582
- PMCID: PMC3266594
- DOI: 10.1128/MCB.06455-11
Conserved molecular interactions within the HBO1 acetyltransferase complexes regulate cell proliferation
Abstract
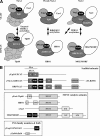
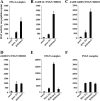
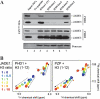
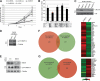
Acetyltransferase complexes of the MYST family with distinct substrate specificities and functions maintain a conserved association with different ING tumor suppressor proteins. ING complexes containing the HBO1 acetylase are a major source of histone H3 and H4 acetylation in vivo and play critical roles in gene regulation and DNA replication. Here, our molecular dissection of HBO1/ING complexes unravels the protein domains required for their assembly and function. Multiple PHD finger domains present in different subunits bind the histone H3 N-terminal tail with a distinct specificity toward lysine 4 methylation status. We show that natively regulated association of the ING4/5 PHD domain with HBO1-JADE determines the growth inhibitory function of the complex, linked to its tumor suppressor activity. Functional genomic analyses indicate that the p53 pathway is a main target of the complex, at least in part through direct transcription regulation at the initiation site of p21/CDKN1A. These results demonstrate the importance of ING association with MYST acetyltransferases in controlling cell proliferation, a regulated link that accounts for the reported tumor suppressor activities of these complexes.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
