Eph receptor function is modulated by heterooligomerization of A and B type Eph receptors
- PMID: 22144690
- PMCID: PMC3241718
- DOI: 10.1083/jcb.201104037
Eph receptor function is modulated by heterooligomerization of A and B type Eph receptors
Abstract
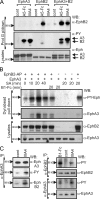
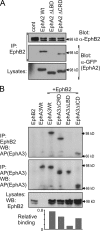
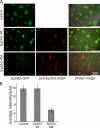
Eph receptors interact with ephrin ligands on adjacent cells to facilitate tissue patterning during normal and oncogenic development, in which unscheduled expression and somatic mutations contribute to tumor progression. EphA and B subtypes preferentially bind A- and B-type ephrins, respectively, resulting in receptor complexes that propagate via homotypic Eph-Eph interactions. We now show that EphA and B receptors cocluster, such that specific ligation of one receptor promotes recruitment and cross-activation of the other. Remarkably, coexpression of a kinase-inactive mutant EphA3 with wild-type EphB2 can cause either cross-activation or cross-inhibition, depending on relative expression. Our findings indicate that cellular responses to ephrin contact are determined by the EphA/EphB receptor profile on a given cell rather than the individual Eph subclass. Importantly, they imply that in tumor cells coexpressing different Ephs, functional mutations in one subtype may cause phenotypes that are a result of altered signaling from heterotypic rather from homotypic Eph clusters.
Figures




References
-
- Alonso-C L.M., Trinidad E.M., de Garcillan B., Ballesteros M., Castellanos M., Cotillo I., Muñoz J.J., Zapata A.G. 2009. Expression profile of Eph receptors and ephrin ligands in healthy human B lymphocytes and chronic lymphocytic leukemia B-cells. Leuk. Res. 33:395–406 10.1016/j.leukres.2008.08.010 - DOI - PubMed
-
- Binns K.L., Taylor P.P., Sicheri F., Pawson T., Holland S.J. 2000. Phosphorylation of tyrosine residues in the kinase domain and juxtamembrane region regulates the biological and catalytic activities of Eph receptors. Mol. Cell. Biol. 20:4791–4805 10.1128/MCB.20.13.4791-4805.2000 - DOI - PMC - PubMed
-
- Brown A., Yates P.A., Burrola P., Ortuño D., Vaidya A., Jessell T.M., Pfaff S.L., O’Leary D.D., Lemke G. 2000. Topographic mapping from the retina to the midbrain is controlled by relative but not absolute levels of EphA receptor signaling. Cell. 102:77–88 10.1016/S0092-8674(00)00012-X - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

