Comparative microbial modules resource: generation and visualization of multi-species biclusters
- PMID: 22144874
- PMCID: PMC3228777
- DOI: 10.1371/journal.pcbi.1002228
Comparative microbial modules resource: generation and visualization of multi-species biclusters
Abstract
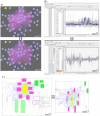
The increasing abundance of large-scale, high-throughput datasets for many closely related organisms provides opportunities for comparative analysis via the simultaneous biclustering of datasets from multiple species. These analyses require a reformulation of how to organize multi-species datasets and visualize comparative genomics data analyses results. Recently, we developed a method, multi-species cMonkey, which integrates heterogeneous high-throughput datatypes from multiple species to identify conserved regulatory modules. Here we present an integrated data visualization system, built upon the Gaggle, enabling exploration of our method's results (available at http://meatwad.bio.nyu.edu/cmmr.html). The system can also be used to explore other comparative genomics datasets and outputs from other data analysis procedures - results from other multiple-species clustering programs or from independent clustering of different single-species datasets. We provide an example use of our system for two bacteria, Escherichia coli and Salmonella Typhimurium. We illustrate the use of our system by exploring conserved biclusters involved in nitrogen metabolism, uncovering a putative function for yjjI, a currently uncharacterized gene that we predict to be involved in nitrogen assimilation.
© 2011 Kacmarczyk et al.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

