Validation of skeletal muscle cis-regulatory module predictions reveals nucleotide composition bias in functional enhancers
- PMID: 22144875
- PMCID: PMC3228787
- DOI: 10.1371/journal.pcbi.1002256
Validation of skeletal muscle cis-regulatory module predictions reveals nucleotide composition bias in functional enhancers
Abstract
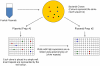
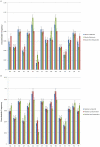
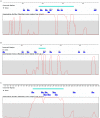
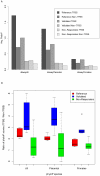
We performed a genome-wide scan for muscle-specific cis-regulatory modules (CRMs) using three computational prediction programs. Based on the predictions, 339 candidate CRMs were tested in cell culture with NIH3T3 fibroblasts and C2C12 myoblasts for capacity to direct selective reporter gene expression to differentiated C2C12 myotubes. A subset of 19 CRMs validated as functional in the assay. The rate of predictive success reveals striking limitations of computational regulatory sequence analysis methods for CRM discovery. Motif-based methods performed no better than predictions based only on sequence conservation. Analysis of the properties of the functional sequences relative to inactive sequences identifies nucleotide sequence composition can be an important characteristic to incorporate in future methods for improved predictive specificity. Muscle-related TFBSs predicted within the functional sequences display greater sequence conservation than non-TFBS flanking regions. Comparison with recent MyoD and histone modification ChIP-Seq data supports the validity of the functional regions.
© 2011 Kwon et al.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Arnone MI, Davidson EH. The hardwiring of development: organization and function of genomic regulatory systems. Development. 1997;124:1851–1864. - PubMed
-
- Shimizu S, Miyamoto Y, Hayashi M. Cell-type dependency of two Foxa/HNF3 sites in the regulation of vitronectin promoter activity. Biochim Biophys Acta. 2002;1574:337–344. - PubMed
-
- McBride DJ, Kleinjan DA. Rounding up active cis-elements in the triple C corral: combining conservation, cleavage and conformation capture for the analysis of regulatory gene domains. Brief Funct Genomic Proteomic. 2004;3:267–279. - PubMed
-
- Ling J, Ainol L, Zhang L, Yu X, Pi W, et al. HS2 enhancer function is blocked by a transcriptional terminator inserted between the enhancer and the promoter. J Biol Chem. 2004;279:51704–51713. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

