Estimated comparative integration hotspots identify different behaviors of retroviral gene transfer vectors
- PMID: 22144885
- PMCID: PMC3228801
- DOI: 10.1371/journal.pcbi.1002292
Estimated comparative integration hotspots identify different behaviors of retroviral gene transfer vectors
Abstract
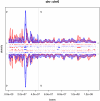
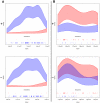
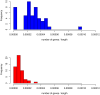
Integration of retroviral vectors in the human genome follows non random patterns that favor insertional deregulation of gene expression and may cause risks of insertional mutagenesis when used in clinical gene therapy. Understanding how viral vectors integrate into the human genome is a key issue in predicting these risks. We provide a new statistical method to compare retroviral integration patterns. We identified the positions where vectors derived from the Human Immunodeficiency Virus (HIV) and the Moloney Murine Leukemia Virus (MLV) show different integration behaviors in human hematopoietic progenitor cells. Non-parametric density estimation was used to identify candidate comparative hotspots, which were then tested and ranked. We found 100 significative comparative hotspots, distributed throughout the chromosomes. HIV hotspots were wider and contained more genes than MLV ones. A Gene Ontology analysis of HIV targets showed enrichment of genes involved in antigen processing and presentation, reflecting the high HIV integration frequency observed at the MHC locus on chromosome 6. Four histone modifications/variants had a different mean density in comparative hotspots (H2AZ, H3K4me1, H3K4me3, H3K9me1), while gene expression within the comparative hotspots did not differ from background. These findings suggest the existence of epigenetic or nuclear three-dimensional topology contexts guiding retroviral integration to specific chromosome areas.
© 2011 Ambrosi et al.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Deletion of the LTR enhancer/promoter has no impact on the integration profile of MLV vectors in human hematopoietic progenitors.PLoS One. 2013;8(1):e55721. doi: 10.1371/journal.pone.0055721. Epub 2013 Jan 31. PLoS One. 2013. PMID: 23383272 Free PMC article.
-
High-definition mapping of retroviral integration sites identifies active regulatory elements in human multipotent hematopoietic progenitors.Blood. 2010 Dec 16;116(25):5507-17. doi: 10.1182/blood-2010-05-283523. Epub 2010 Sep 23. Blood. 2010. PMID: 20864581
-
Enhancers are major targets for murine leukemia virus vector integration.J Virol. 2014 Apr;88(8):4504-13. doi: 10.1128/JVI.00011-14. Epub 2014 Feb 5. J Virol. 2014. PMID: 24501411 Free PMC article.
-
Stem cell gene transfer: insights into integration and hematopoiesis from primate genetic marking studies.Ann N Y Acad Sci. 2005 Jun;1044:178-82. doi: 10.1196/annals.1349.023. Ann N Y Acad Sci. 2005. PMID: 15958711 Review.
-
Determinants of Retroviral Integration and Implications for Gene Therapeutic MLV-Based Vectors and for a Cure for HIV-1 Infection.Viruses. 2022 Dec 21;15(1):32. doi: 10.3390/v15010032. Viruses. 2022. PMID: 36680071 Free PMC article. Review.
Cited by
-
Genome-wide analysis of alpharetroviral integration in human hematopoietic stem/progenitor cells.Genes (Basel). 2014 May 16;5(2):415-29. doi: 10.3390/genes5020415. Genes (Basel). 2014. PMID: 24840152 Free PMC article.
-
Sleeping Beauty-Mediated Drug Resistance Gene Transfer in Human Hematopoietic Progenitor Cells.Hum Gene Ther. 2015 Oct;26(10):657-63. doi: 10.1089/hum.2015.058. Epub 2015 Sep 18. Hum Gene Ther. 2015. PMID: 26176276 Free PMC article.
-
Methylome-wide Analysis of Chronic HIV Infection Reveals Five-Year Increase in Biological Age and Epigenetic Targeting of HLA.Mol Cell. 2016 Apr 21;62(2):157-168. doi: 10.1016/j.molcel.2016.03.019. Mol Cell. 2016. PMID: 27105112 Free PMC article.
-
Lentivector integration sites in ependymal cells from a model of metachromatic leukodystrophy: non-B DNA as a new factor influencing integration.Mol Ther Nucleic Acids. 2014 Aug 26;3(8):e187. doi: 10.1038/mtna.2014.39. Mol Ther Nucleic Acids. 2014. PMID: 25158091 Free PMC article.
-
Progresses towards safe and efficient gene therapy vectors.Oncotarget. 2015 Oct 13;6(31):30675-703. doi: 10.18632/oncotarget.5169. Oncotarget. 2015. PMID: 26362400 Free PMC article. Review.
References
-
- Aiuti A, Cattaneo F, Galimberti S, Benninghoff U, Cassani B, et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med. 2009;360:447–458. - PubMed
-
- Hacein-Bey-Abina S, Le Deist F, Carlier F, Bouneaud C, Hue C, et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;346:1185–1193. - PubMed
-
- Mavilio F, Pellegrini G, Ferrari S, Di Nunzio F, Di Iorio E, et al. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nat Med. 2006;12:1397–1402. - PubMed
-
- Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

