SREBP coordinates iron and ergosterol homeostasis to mediate triazole drug and hypoxia responses in the human fungal pathogen Aspergillus fumigatus
- PMID: 22144905
- PMCID: PMC3228822
- DOI: 10.1371/journal.pgen.1002374
SREBP coordinates iron and ergosterol homeostasis to mediate triazole drug and hypoxia responses in the human fungal pathogen Aspergillus fumigatus
Abstract
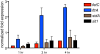
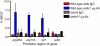
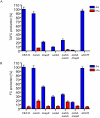
Sterol regulatory element binding proteins (SREBPs) are a class of basic helix-loop-helix transcription factors that regulate diverse cellular responses in eukaryotes. Adding to the recognized importance of SREBPs in human health, SREBPs in the human fungal pathogens Cryptococcus neoformans and Aspergillus fumigatus are required for fungal virulence and susceptibility to triazole antifungal drugs. To date, the exact mechanism(s) behind the role of SREBP in these observed phenotypes is not clear. Here, we report that A. fumigatus SREBP, SrbA, mediates regulation of iron acquisition in response to hypoxia and low iron conditions. To further define SrbA's role in iron acquisition in relation to previously studied fungal regulators of iron metabolism, SreA and HapX, a series of mutants were generated in the ΔsrbA background. These data suggest that SrbA is activated independently of SreA and HapX in response to iron limitation, but that HapX mRNA induction is partially dependent on SrbA. Intriguingly, exogenous addition of high iron or genetic deletion of sreA in the ΔsrbA background was able to partially rescue the hypoxia growth, triazole drug susceptibility, and decrease in ergosterol content phenotypes of ΔsrbA. Thus, we conclude that the fungal SREBP, SrbA, is critical for coordinating genes involved in iron acquisition and ergosterol biosynthesis under hypoxia and low iron conditions found at sites of human fungal infections. These results support a role for SREBP-mediated iron regulation in fungal virulence, and they lay a foundation for further exploration of SREBP's role in iron homeostasis in other eukaryotes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



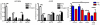


Similar articles
-
Regulation of High-Affinity Iron Acquisition, Including Acquisition Mediated by the Iron Permease FtrA, Is Coordinated by AtrR, SrbA, and SreA in Aspergillus fumigatus.mBio. 2023 Jun 27;14(3):e0075723. doi: 10.1128/mbio.00757-23. Epub 2023 Apr 24. mBio. 2023. PMID: 37093084 Free PMC article.
-
SREBP-dependent triazole susceptibility in Aspergillus fumigatus is mediated through direct transcriptional regulation of erg11A (cyp51A).Antimicrob Agents Chemother. 2012 Jan;56(1):248-57. doi: 10.1128/AAC.05027-11. Epub 2011 Oct 17. Antimicrob Agents Chemother. 2012. PMID: 22006005 Free PMC article.
-
ChIP-seq and in vivo transcriptome analyses of the Aspergillus fumigatus SREBP SrbA reveals a new regulator of the fungal hypoxia response and virulence.PLoS Pathog. 2014 Nov 6;10(11):e1004487. doi: 10.1371/journal.ppat.1004487. eCollection 2014 Nov. PLoS Pathog. 2014. PMID: 25375670 Free PMC article.
-
Fungal iron homeostasis with a focus on Aspergillus fumigatus.Biochim Biophys Acta Mol Cell Res. 2021 Jan;1868(1):118885. doi: 10.1016/j.bbamcr.2020.118885. Epub 2020 Oct 10. Biochim Biophys Acta Mol Cell Res. 2021. PMID: 33045305 Review.
-
Regulation of Sterol Biosynthesis in the Human Fungal Pathogen Aspergillus fumigatus: Opportunities for Therapeutic Development.Front Microbiol. 2017 Feb 1;8:92. doi: 10.3389/fmicb.2017.00092. eCollection 2017. Front Microbiol. 2017. PMID: 28203225 Free PMC article. Review.
Cited by
-
Hypoxia: A Double-Edged Sword During Fungal Pathogenesis?Front Microbiol. 2020 Aug 12;11:1920. doi: 10.3389/fmicb.2020.01920. eCollection 2020. Front Microbiol. 2020. PMID: 32903454 Free PMC article. Review.
-
Mga2 Transcription Factor Regulates an Oxygen-responsive Lipid Homeostasis Pathway in Fission Yeast.J Biol Chem. 2016 Jun 3;291(23):12171-83. doi: 10.1074/jbc.M116.723650. Epub 2016 Apr 6. J Biol Chem. 2016. PMID: 27053105 Free PMC article.
-
Online Biomass Monitoring Enables Characterization of the Growth Pattern of Aspergillus fumigatus in Liquid Shake Conditions.J Fungi (Basel). 2022 Sep 27;8(10):1013. doi: 10.3390/jof8101013. J Fungi (Basel). 2022. PMID: 36294578 Free PMC article.
-
Transcriptional Control of Drug Resistance, Virulence and Immune System Evasion in Pathogenic Fungi: A Cross-Species Comparison.Front Cell Infect Microbiol. 2016 Oct 20;6:131. doi: 10.3389/fcimb.2016.00131. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 27812511 Free PMC article. Review.
-
The Transcription Factor VdHapX Controls Iron Homeostasis and Is Crucial for Virulence in the Vascular Pathogen Verticillium dahliae.mSphere. 2018 Sep 5;3(5):e00400-18. doi: 10.1128/mSphere.00400-18. mSphere. 2018. PMID: 30185514 Free PMC article.
References
-
- Kornitzer D. Fungal mechanisms for host iron acquisition. Current opinion in microbiology. 2009;12:377–383. - PubMed
-
- Schrettl M, Beckmann N, Varga J, Heinekamp T, Jacobsen ID, et al. HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog. 2010;6:e1001124. doi: 10.1371/journal.ppat.1001124. - DOI - PMC - PubMed
-
- Schrettl M, Bignell E, Kragl C, Sabiha Y, Loss O, et al. Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog. 2007;3:e128. doi: 10.1371/journal.ppat.0030128. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

