Repetitive elements may comprise over two-thirds of the human genome
- PMID: 22144907
- PMCID: PMC3228813
- DOI: 10.1371/journal.pgen.1002384
Repetitive elements may comprise over two-thirds of the human genome
Abstract
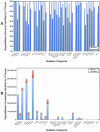
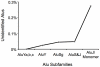
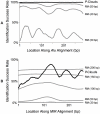
Transposable elements (TEs) are conventionally identified in eukaryotic genomes by alignment to consensus element sequences. Using this approach, about half of the human genome has been previously identified as TEs and low-complexity repeats. We recently developed a highly sensitive alternative de novo strategy, P-clouds, that instead searches for clusters of high-abundance oligonucleotides that are related in sequence space (oligo "clouds"). We show here that P-clouds predicts >840 Mbp of additional repetitive sequences in the human genome, thus suggesting that 66%-69% of the human genome is repetitive or repeat-derived. To investigate this remarkable difference, we conducted detailed analyses of the ability of both P-clouds and a commonly used conventional approach, RepeatMasker (RM), to detect different sized fragments of the highly abundant human Alu and MIR SINEs. RM can have surprisingly low sensitivity for even moderately long fragments, in contrast to P-clouds, which has good sensitivity down to small fragment sizes (∼25 bp). Although short fragments have a high intrinsic probability of being false positives, we performed a probabilistic annotation that reflects this fact. We further developed "element-specific" P-clouds (ESPs) to identify novel Alu and MIR SINE elements, and using it we identified ∼100 Mb of previously unannotated human elements. ESP estimates of new MIR sequences are in good agreement with RM-based predictions of the amount that RM missed. These results highlight the need for combined, probabilistic genome annotation approaches and suggest that the human genome consists of substantially more repetitive sequence than previously believed.
Figures



References
-
- Frith MC, Pheasant M, Mattick JS. Genomics: The amazing complexity of the human transcriptome. Eur J Hum Genet. 2005;13:894–897. - PubMed
-
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–29. - PubMed
-
- Pheasant M, Mattick JS. Raising the estimate of functional human sequences. Genome Res. 2007;17:1245–1253. - PubMed
-
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. - PubMed
-
- Eichler EE. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 2001;17:661–669. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

