A population genetics-phylogenetics approach to inferring natural selection in coding sequences
- PMID: 22144911
- PMCID: PMC3228810
- DOI: 10.1371/journal.pgen.1002395
A population genetics-phylogenetics approach to inferring natural selection in coding sequences
Abstract
Through an analysis of polymorphism within and divergence between species, we can hope to learn about the distribution of selective effects of mutations in the genome, changes in the fitness landscape that occur over time, and the location of sites involved in key adaptations that distinguish modern-day species. We introduce a novel method for the analysis of variation in selection pressures within and between species, spatially along the genome and temporally between lineages. We model codon evolution explicitly using a joint population genetics-phylogenetics approach that we developed for the construction of multiallelic models with mutation, selection, and drift. Our approach has the advantage of performing direct inference on coding sequences, inferring ancestral states probabilistically, utilizing allele frequency information, and generalizing to multiple species. We use a Bayesian sliding window model for intragenic variation in selection coefficients that efficiently combines information across sites and captures spatial clustering within the genome. To demonstrate the utility of the method, we infer selective pressures acting in Drosophila melanogaster and D. simulans from polymorphism and divergence data for 100 X-linked coding regions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
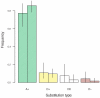
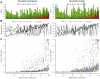
 : mean selection coefficient at viable sites.
: mean selection coefficient at viable sites.  : proportion of sites viable.
: proportion of sites viable.  : odds ratio of the McDonald-Kreitman table. (C)
: odds ratio of the McDonald-Kreitman table. (C)  , the posterior probability of positive selection per codon (points) and per gene (black line). Points are colored randomly to aid visualization. In (A), (B) and (C), genes are ordered horizontally by the rank of
, the posterior probability of positive selection per codon (points) and per gene (black line). Points are colored randomly to aid visualization. In (A), (B) and (C), genes are ordered horizontally by the rank of  per gene.
per gene.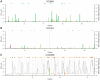
Similar articles
-
Mutation and selection at silent and replacement sites in the evolution of animal mitochondrial DNA.Genetica. 1998;102-103(1-6):393-407. Genetica. 1998. PMID: 9720291
-
The evolution of small insertions and deletions in the coding genes of Drosophila melanogaster.Mol Biol Evol. 2013 Dec;30(12):2699-708. doi: 10.1093/molbev/mst167. Epub 2013 Sep 26. Mol Biol Evol. 2013. PMID: 24077769
-
Inferring weak selection from patterns of polymorphism and divergence at "silent" sites in Drosophila DNA.Genetics. 1995 Feb;139(2):1067-76. doi: 10.1093/genetics/139.2.1067. Genetics. 1995. PMID: 7713409 Free PMC article.
-
Patterns of polymorphism and divergence from noncoding sequences of Drosophila melanogaster and D. simulans: evidence for nonequilibrium processes.Mol Biol Evol. 2005 Jan;22(1):51-62. doi: 10.1093/molbev/msh269. Epub 2004 Sep 29. Mol Biol Evol. 2005. PMID: 15456897 Review.
-
Codon bias evolution in Drosophila. Population genetics of mutation-selection drift.Gene. 1997 Dec 31;205(1-2):269-78. doi: 10.1016/s0378-1119(97)00400-9. Gene. 1997. PMID: 9461401 Review.
Cited by
-
Inferences of demography and selection in an African population of Drosophila melanogaster.Genetics. 2013 Jan;193(1):215-28. doi: 10.1534/genetics.112.145318. Epub 2012 Oct 26. Genetics. 2013. PMID: 23105013 Free PMC article.
-
Genetic conflicts with Plasmodium parasites and functional constraints shape the evolution of erythrocyte cytoskeletal proteins.Sci Rep. 2018 Oct 2;8(1):14682. doi: 10.1038/s41598-018-33049-y. Sci Rep. 2018. PMID: 30279439 Free PMC article.
-
Discovering recent selection forces shaping the evolution of dengue viruses based on polymorphism data across geographic scales.Virus Evol. 2022 Nov 29;8(2):veac108. doi: 10.1093/ve/veac108. eCollection 2022. Virus Evol. 2022. PMID: 36601300 Free PMC article.
-
Comparative population genomics: power and principles for the inference of functionality.Trends Genet. 2014 Apr;30(4):133-9. doi: 10.1016/j.tig.2014.02.002. Epub 2014 Mar 20. Trends Genet. 2014. PMID: 24656563 Free PMC article. Review.
-
An evolutionary analysis of antigen processing and presentation across different timescales reveals pervasive selection.PLoS Genet. 2014 Mar 27;10(3):e1004189. doi: 10.1371/journal.pgen.1004189. eCollection 2014 Mar. PLoS Genet. 2014. PMID: 24675550 Free PMC article.
References
-
- Tinbergen N. On aims and methods of ethology. Zeitschrift für Tierpsychologie. 1963;20:410–433.
-
- Gould SJ, Lewontin RC. The spandrels of San Marco and the Panglossian paradigm. Proc Roy Soc Lond B. 1979;205:581–598. - PubMed
-
- Kimura M. The Neutral Theory of Molecular Evolution. 1983. Cambridge University Press, Cambridge. - PubMed
-
- Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nat Rev Genet. 2007;8:610–618. - PubMed
-
- Sella G, Petrov D, Przeworski M, Andolfatto P. Pervasive natural selection in the Drosophila genome? PLoS Genet. 2009;5:e1000495. doi: 10.1371/journal.pgen.1000495. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

