Transcriptome analysis of Stagonospora nodorum: gene models, effectors, metabolism and pantothenate dispensability
- PMID: 22145589
- PMCID: PMC6638697
- DOI: 10.1111/j.1364-3703.2011.00770.x
Transcriptome analysis of Stagonospora nodorum: gene models, effectors, metabolism and pantothenate dispensability
Abstract
The wheat pathogen Stagonospora nodorum, causal organism of the wheat disease Stagonospora nodorum blotch, has emerged as a model for the Dothideomycetes, a large fungal taxon that includes many important plant pathogens. The initial annotation of the genome assembly included 16,586 nuclear gene models. These gene models were used to design a microarray that has been interrogated with labelled transcripts from six cDNA samples: four from infected wheat plants at time points spanning early infection to sporulation, and two time points taken from growth in artificial media. Positive signals of expression were obtained for 12,281 genes. This represents strong corroborative evidence of the validity of these gene models. Significantly differential expression between the various time points was observed. When infected samples were compared with axenic cultures, 2882 genes were expressed at a higher level in planta and 3630 were expressed more highly in vitro. Similar numbers were differentially expressed between different developmental stages. The earliest time points in planta were particularly enriched in differentially expressed genes. A disproportionate number of the early expressed gene products were predicted to be secreted, but otherwise had no obvious sequence homology to functionally characterized genes. These genes are candidate necrotrophic effectors. We have focused attention on genes for carbohydrate metabolism and the specific biosynthetic pathways active during growth in planta. The analysis points to a very dynamic adjustment of metabolism during infection. Functional analysis of a gene in the coenzyme A biosynthetic pathway showed that the enzyme was dispensable for growth, indicating that a precursor is supplied by the plant.
© 2011 The Authors. Molecular Plant Pathology © 2011 BSPP and Blackwell Publishing Ltd.
Figures

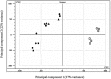
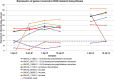
 ), 5 dpi in planta (◆), 7 dpi in planta (●) and 10 dpi in planta (▴)] and axenically grown SN15 [4 dpi in vitro (
), 5 dpi in planta (◆), 7 dpi in planta (●) and 10 dpi in planta (▴)] and axenically grown SN15 [4 dpi in vitro ( ) and 16 dpi in vitro (
) and 16 dpi in vitro ( )]. Principal component 1 (PC1) accounted for 37% of the variation and principal component 2 (PC2) described 22% of the variation. PC1 mainly differentiates samples according to the growth medium, whereas PC2 mainly differentiates samples according to their developmental stage. dpi, days post‐inoculation.
)]. Principal component 1 (PC1) accounted for 37% of the variation and principal component 2 (PC2) described 22% of the variation. PC1 mainly differentiates samples according to the growth medium, whereas PC2 mainly differentiates samples according to their developmental stage. dpi, days post‐inoculation.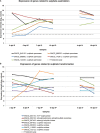
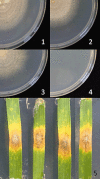


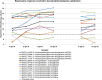
Similar articles
-
Dothideomycete plant interactions illuminated by genome sequencing and EST analysis of the wheat pathogen Stagonospora nodorum.Plant Cell. 2007 Nov;19(11):3347-68. doi: 10.1105/tpc.107.052829. Epub 2007 Nov 16. Plant Cell. 2007. PMID: 18024570 Free PMC article.
-
Genetics of Variable Disease Expression Conferred by Inverse Gene-For-Gene Interactions in the Wheat-Parastagonospora nodorum Pathosystem.Plant Physiol. 2019 May;180(1):420-434. doi: 10.1104/pp.19.00149. Epub 2019 Mar 11. Plant Physiol. 2019. PMID: 30858234 Free PMC article.
-
Stagonospora nodorum: from pathology to genomics and host resistance.Annu Rev Phytopathol. 2012;50:23-43. doi: 10.1146/annurev-phyto-081211-173019. Epub 2012 May 1. Annu Rev Phytopathol. 2012. PMID: 22559071 Review.
-
Proteomic identification of extracellular proteins regulated by the Gna1 Galpha subunit in Stagonospora nodorum.Mycol Res. 2009 May;113(5):523-31. doi: 10.1016/j.mycres.2009.01.004. Epub 2009 Jan 22. Mycol Res. 2009. PMID: 19284980
-
Biology and molecular interactions of Parastagonospora nodorum blotch of wheat.Planta. 2021 Dec 16;255(1):21. doi: 10.1007/s00425-021-03796-w. Planta. 2021. PMID: 34914013 Review.
Cited by
-
Strategies for Wheat Stripe Rust Pathogenicity Identified by Transcriptome Sequencing.PLoS One. 2013 Jun 26;8(6):e67150. doi: 10.1371/journal.pone.0067150. Print 2013. PLoS One. 2013. PMID: 23840606 Free PMC article.
-
Genomes and transcriptomes of partners in plant-fungal-interactions between canola (Brassica napus) and two Leptosphaeria species.PLoS One. 2014 Jul 28;9(7):e103098. doi: 10.1371/journal.pone.0103098. eCollection 2014. PLoS One. 2014. PMID: 25068644 Free PMC article.
-
Chromosome-level genome assembly and manually-curated proteome of model necrotroph Parastagonospora nodorum Sn15 reveals a genome-wide trove of candidate effector homologs, and redundancy of virulence-related functions within an accessory chromosome.BMC Genomics. 2021 May 25;22(1):382. doi: 10.1186/s12864-021-07699-8. BMC Genomics. 2021. PMID: 34034667 Free PMC article.
-
The Velvet transcription factor PnVeA regulates necrotrophic effectors and secondary metabolism in the wheat pathogen Parastagonospora nodorum.BMC Microbiol. 2024 Aug 10;24(1):299. doi: 10.1186/s12866-024-03454-7. BMC Microbiol. 2024. PMID: 39127645 Free PMC article.
-
Differential effector gene expression underpins epistasis in a plant fungal disease.Plant J. 2016 Aug;87(4):343-54. doi: 10.1111/tpj.13203. Epub 2016 Jul 7. Plant J. 2016. PMID: 27133896 Free PMC article.
References
-
- Andersen, B. , Dongo, A. and Pryor, B.M. (2008) Secondary metabolite profiling of Alternaria dauci, A. porri, A. solani, and A. tomatophila . Mycol. Res. 112, 241–250. - PubMed
-
- Bailey, A. , Mueller, E. and Bowyer, P. (2000) Ornithine decarboxylase of Stagonospora (Septoria) nodorum is required for virulence toward wheat. J. Biol. Chem. 275, 14 242–14 247. - PubMed
-
- Benjamini, Y. and Hochberg, Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 57, 289–300.
-
- Blixt, E. , Djurle, A. , Yuen, J. and Olson, Ã. (2009) Fungicide sensitivity in Swedish isolates of Phaeosphaeria nodorum . Plant Pathol. 58, 655–664.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

