Inhibition of HIV-1 infection by TNPO3 depletion is determined by capsid and detectable after viral cDNA enters the nucleus
- PMID: 22145813
- PMCID: PMC3267670
- DOI: 10.1186/1742-4690-8-98
Inhibition of HIV-1 infection by TNPO3 depletion is determined by capsid and detectable after viral cDNA enters the nucleus
Abstract
Background: HIV-1 infects non-dividing cells. This implies that the virus traverses the nuclear pore before it integrates into chromosomal DNA. Recent studies demonstrated that TNPO3 is required for full infectivity of HIV-1. The fact that TNPO3 is a karyopherin suggests that it acts by directly promoting nuclear entry of HIV-1. Some studies support this hypothesis, while others have failed to do so. Additionally, some studies suggest that TNPO3 acts via HIV-1 Integrase (IN), and others indicate that it acts via capsid (CA).
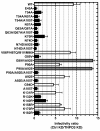
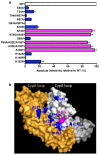
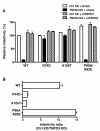
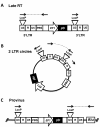
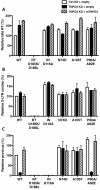
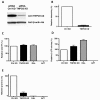
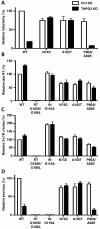
Results: To shed light on the mechanism by which TNPO3 contributes to HIV-1 infection we engineered a panel of twenty-seven single-cycle HIV-1 vectors each bearing a different CA mutation and characterized them for the ability to transduce cells in which TNPO3 had been knocked down (KD). Fourteen CA mutants were relatively TNPO3-independent, as compared to wild-type (WT) HIV-1. Two mutants were more TNPO3-dependent than the WT, and eleven mutants were actually inhibited by TNPO3. The efficiency of the synthesis of viral cDNA, 2-LTR circles, and proviral DNA was then assessed for WT HIV-1 and three select CA mutants. Controls included rescue of TNPO3 KD with non-targetable coding sequence, RT- and IN- mutant viruses, and pharmacologic inhibitors of RT and IN. TNPO3 KD blocked transduction and establishment of proviral DNA by wild-type HIV-1 with no significant effect on the level of 2-LTR circles. PCR results were confirmed by achieving TNPO3 KD using two different methodologies (lentiviral vector and siRNA oligonucleotide transfection); by challenging three different cell types; by using two different challenge viruses, each necessitating different sets of PCR primers; and by pseudotyping virus with VSV G or using HIV-1 Env.
Conclusion: TNPO3 promotes HIV-1 infectivity at a step in the virus life cycle that is detectable after the preintegration complex arrives in the nucleus and CA is the viral determinant for TNPO3 dependence.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

