Temporal competition between differentiation programs determines cell fate choice
- PMID: 22146301
- PMCID: PMC3737729
- DOI: 10.1038/msb.2011.88
Temporal competition between differentiation programs determines cell fate choice
Abstract
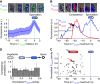
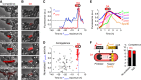
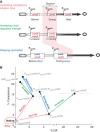
Multipotent differentiation, where cells adopt one of several possible fates, occurs in diverse systems ranging from bacteria to mammals. This decision-making process is driven by multiple differentiation programs that operate simultaneously in the cell. How these programs interact to govern cell fate choice is poorly understood. To investigate this issue, we simultaneously measured activities of the competing sporulation and competence programs in single Bacillus subtilis cells. This approach revealed that these competing differentiation programs progress independently without cross-regulation before the decision point. Cells seem to arrive at a fate choice through differences in the relative timing between the two programs. To test this proposed dynamic mechanism, we altered the relative timing by engineering artificial cross-regulation between the sporulation and competence circuits. Results suggest a simple model that does not require a checkpoint or intricate cross-regulation before cellular decision-making. Rather, cell fate choice appears to be the outcome of a 'molecular race' between differentiation programs that compete in time, providing a simple dynamic mechanism for decision-making.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Comment in
-
Winner takes all in a race for cell fate.Mol Syst Biol. 2011 Dec 6;7:558. doi: 10.1038/msb.2011.92. Mol Syst Biol. 2011. PMID: 22146302 Free PMC article. No abstract available.
-
Differentiation: Racing to decide.Nat Rev Genet. 2011 Dec 20;13(2):72. doi: 10.1038/nrg3158. Nat Rev Genet. 2011. PMID: 22183154 No abstract available.
-
Bacterial development: Racing to decide.Nat Rev Microbiol. 2012 Jan 16;10(2):84. doi: 10.1038/nrmicro2735. Nat Rev Microbiol. 2012. PMID: 22245931 No abstract available.
References
-
- Asayama M, Kobayashi Y (1993) Signal transduction and sporulation in Bacillus subtilis: heterologous phosphorylation of Spo0A, a sporulation initiation gene product. J Biochem 114: 385–388 - PubMed
-
- Bai U, Mandic-Mulec I, Smith I (1993) SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev 7: 139–148 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

