Sample pretreatment and nucleic acid-based detection for fast diagnosis utilizing microfluidic systems
- PMID: 22146901
- PMCID: PMC7088154
- DOI: 10.1007/s10439-011-0473-4
Sample pretreatment and nucleic acid-based detection for fast diagnosis utilizing microfluidic systems
Abstract
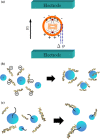
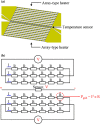
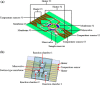
Recently, micro-electro-mechanical-systems (MEMS) technology and micromachining techniques have enabled miniaturization of biomedical devices and systems. Not only do these techniques facilitate the development of miniaturized instrumentation for biomedical analysis, but they also open a new era for integration of microdevices for performing accurate and sensitive diagnostic assays. A so-called "micro-total-analysis-system", which integrates sample pretreatment, transport, reaction, and detection on a small chip in an automatic format, can be realized by combining functional microfluidic components manufactured by specific MEMS technologies. Among the promising applications using microfluidic technologies, nucleic acid-based detection has shown considerable potential recently. For instance, micro-polymerase chain reaction chips for rapid DNA amplification have attracted considerable interest. In addition, microfluidic devices for rapid sample pretreatment prior to nucleic acid-based detection have also achieved significant progress in the recent years. In this review paper, microfluidic systems for sample preparation, nucleic acid amplification and detection for fast diagnosis will be reviewed. These microfluidic devices and systems have several advantages over their large-scale counterparts, including lower sample/reagent consumption, lower power consumption, compact size, faster analysis, and lower per unit cost. The development of these microfluidic devices and systems may provide a revolutionary platform technology for fast sample pretreatment and accurate, sensitive diagnosis.
Figures

References
-
- Ahn CH, et al. A fully integrated micromachined magnetic particle separator. J. Microelectromech. Syst. 1996;5(3):151–158. doi: 10.1109/84.536621. - DOI
-
- Bao N, Lu C. A microfluidic device for physical trapping and electrical lysis of bacterial cells. Appl. Phys. Lett. 2008;92(21):21403. doi: 10.1063/1.2937088. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

