Genome-wide SNP genotyping identifies the Stereocilin (STRC) gene as a major contributor to pediatric bilateral sensorineural hearing impairment
- PMID: 22147502
- PMCID: PMC3264741
- DOI: 10.1002/ajmg.a.34391
Genome-wide SNP genotyping identifies the Stereocilin (STRC) gene as a major contributor to pediatric bilateral sensorineural hearing impairment
Abstract
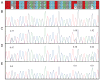
Hearing loss is the most prevalent sensory perception deficit in humans, affecting 1/500 newborns, can be syndromic or nonsyndromic and is genetically heterogeneous. Nearly 80% of inherited nonsyndromic bilateral sensorineural hearing loss (NBSNHI) is autosomal recessive. Although many causal genes have been identified, most are minor contributors, except for GJB2, which accounts for nearly 50% of all recessive cases of severe to profound congenital NBSNHI in some populations. More than 60% of children with a NBSNHI do not have an identifiable genetic cause. To identify genetic contributors, we genotyped 659 GJB2 mutation negative pediatric probands with NBSNHI and assayed for copy number variants (CNVs). After identifying 8 mild-moderate NBSNHI probands with a Chr15q15.3 deletion encompassing the Stereocilin (STRC) gene amongst this cohort, sequencing of STRC was undertaken in these probands as well as 50 probands and 14 siblings with mild-moderate NBSNHI and 40 probands with moderately severe-profound NBSNHI who were GJB2 mutation negative. The existence of a STRC pseudogene that is 99.6% homologous to the STRC coding region has made the sequencing interpretation complicated. We identified 7/50 probands in the mild-moderate cohort to have biallelic alterations in STRC, not including the 8 previously identified deletions. We also identified 2/40 probands to have biallelic alterations in the moderately severe-profound NBSNHI cohort, notably no large deletions in combination with another variant were found in this cohort. The data suggest that STRC may be a common contributor to NBSNHI among GJB2 mutation negative probands, especially in those with mild to moderate hearing impairment.
Copyright © 2011 Wiley Periodicals, Inc.
Figures

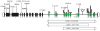
References
-
- Avidan N, Tamary H, Dgany O, Cattan D, Pariente A, Thulliez M, Borot N, Moati L, Barthelme A, Shalmon L, Krasnov T, Ben-Asher E, Olender T, Khen M, Yaniv I, Zaizov R, Shalev H, Delaunay J, Fellous M, Lancet D, Beckmann J. CATSPER2, a human autosomal nonsyndromic male infertility gene. Eur J Hum Genet. 2003;11(7):497–502. - PubMed
-
- Campbell DA, McHale DP, Brown KA, Moynihan LM, Houseman M, Karbani G, Parry G, Janjua AH, Newton V, al-Gazali L, Marksham AF, Lench NJ, Mueller RF. A new locus for non-syndromal, autosomal recessive, sensorineural hearing loss (DFNB16) maps to human chromosome 15q21-q22. J Med Genet. 1997;34(12):1015–1017. - PMC - PubMed
-
- Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38(1):75–81. - PubMed
-
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006a;7(2):85–97. - PubMed
-
- Feuk L, Marshall CR, Wintle RF, Scherer SW. Structural variants: changing the landscape of chromosomes and design of disease studies. Hum Mol Genet. 2006b;15(Spec No 1):R57–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

