MES16, a member of the methylesterase protein family, specifically demethylates fluorescent chlorophyll catabolites during chlorophyll breakdown in Arabidopsis
- PMID: 22147518
- PMCID: PMC3271755
- DOI: 10.1104/pp.111.188870
MES16, a member of the methylesterase protein family, specifically demethylates fluorescent chlorophyll catabolites during chlorophyll breakdown in Arabidopsis
Abstract
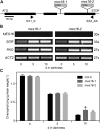
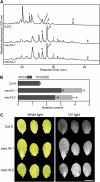
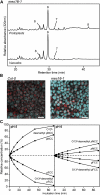
During leaf senescence, chlorophyll (Chl) is broken down to nonfluorescent chlorophyll catabolites (NCCs). These arise from intermediary fluorescent chlorophyll catabolites (FCCs) by an acid-catalyzed isomerization inside the vacuole. The chemical structures of NCCs from Arabidopsis (Arabidopsis thaliana) indicate the presence of an enzyme activity that demethylates the C13(2)-carboxymethyl group present at the isocyclic ring of Chl. Here, we identified this activity as methylesterase family member 16 (MES16; At4g16690). During senescence, mes16 leaves exhibited a strong ultraviolet-excitable fluorescence, which resulted from large amounts of different FCCs accumulating in the mutants. As confirmed by mass spectrometry, these FCCs had an intact carboxymethyl group, which slowed down their isomerization to respective NCCs. Like a homologous protein cloned from radish (Raphanus sativus) and named pheophorbidase, MES16 catalyzed the demethylation of pheophorbide, an early intermediate of Chl breakdown, in vitro, but MES16 also demethylated an FCC. To determine the in vivo substrate of MES16, we analyzed pheophorbide a oxygenase1 (pao1), which is deficient in pheophorbide catabolism and accumulates pheophorbide in the chloroplast, and a mes16pao1 double mutant. In the pao1 background, we additionally mistargeted MES16 to the chloroplast. Normally, MES16 localizes to the cytosol, as shown by analysis of a MES16-green fluorescent protein fusion. Analysis of the accumulating pigments in these lines revealed that pheophorbide is only accessible for demethylation when MES16 is targeted to the chloroplast. Together, these data demonstrate that MES16 is an integral component of Chl breakdown in Arabidopsis and specifically demethylates Chl catabolites at the level of FCCs in the cytosol.
Figures

Similar articles
-
Cytochrome P450 CYP89A9 is involved in the formation of major chlorophyll catabolites during leaf senescence in Arabidopsis.Plant Cell. 2013 May;25(5):1868-80. doi: 10.1105/tpc.113.112151. Epub 2013 May 30. Plant Cell. 2013. PMID: 23723324 Free PMC article.
-
Chlorophyll breakdown in senescent Arabidopsis leaves. Characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction.Plant Physiol. 2005 Sep;139(1):52-63. doi: 10.1104/pp.105.065870. Epub 2005 Aug 19. Plant Physiol. 2005. PMID: 16113212 Free PMC article.
-
Crystal structure of methylesterase family member 16 (MES16) from Arabidopsis thaliana.Biochem Biophys Res Commun. 2016 May 20;474(1):226-231. doi: 10.1016/j.bbrc.2016.04.115. Epub 2016 Apr 22. Biochem Biophys Res Commun. 2016. PMID: 27109476
-
Chlorophyll breakdown in higher plants and algae.Cell Mol Life Sci. 1999 Oct 15;56(3-4):330-47. doi: 10.1007/s000180050434. Cell Mol Life Sci. 1999. PMID: 11212360 Free PMC article. Review.
-
Chlorophyll degradation during senescence.Annu Rev Plant Biol. 2006;57:55-77. doi: 10.1146/annurev.arplant.57.032905.105212. Annu Rev Plant Biol. 2006. PMID: 16669755 Review.
Cited by
-
Structures of chlorophyll catabolites in bananas (Musa acuminata) reveal a split path of chlorophyll breakdown in a ripening fruit.Chemistry. 2012 Aug 27;18(35):10873-85. doi: 10.1002/chem.201201023. Epub 2012 Jul 16. Chemistry. 2012. PMID: 22807397 Free PMC article.
-
Cytochrome P450 CYP89A9 is involved in the formation of major chlorophyll catabolites during leaf senescence in Arabidopsis.Plant Cell. 2013 May;25(5):1868-80. doi: 10.1105/tpc.113.112151. Epub 2013 May 30. Plant Cell. 2013. PMID: 23723324 Free PMC article.
-
A Role for TIC55 as a Hydroxylase of Phyllobilins, the Products of Chlorophyll Breakdown during Plant Senescence.Plant Cell. 2016 Oct;28(10):2510-2527. doi: 10.1105/tpc.16.00630. Epub 2016 Sep 21. Plant Cell. 2016. PMID: 27655840 Free PMC article.
-
Enzyme action in the regulation of plant hormone responses.J Biol Chem. 2013 Jul 5;288(27):19304-11. doi: 10.1074/jbc.R113.475160. Epub 2013 May 24. J Biol Chem. 2013. PMID: 23709222 Free PMC article. Review.
-
Colorless chlorophyll catabolites in senescent florets of broccoli (Brassica oleracea var. italica).J Agric Food Chem. 2015 Feb 11;63(5):1385-92. doi: 10.1021/jf5055326. Epub 2015 Feb 3. J Agric Food Chem. 2015. PMID: 25620234 Free PMC article.
References
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Arnon DI, Whatley FR, Allen MB. (1959) Photosynthesis by isolated chloroplasts. VIII. Photosynthetic phosphorylation and the generation of assimilatory power. Biochim Biophys Acta 32: 47–57 - PubMed
-
- Balzergue S, Dubreucq B, Chauvin S, Le-Clainche I, Le Boulaire F, de Rose R, Samson F, Biaudet V, Lecharny A, Cruaud C, et al. (2001) Improved PCR-walking for large-scale isolation of plant T-DNA borders. Biotechniques 30: 496–498, 502, 504 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

