Next-generation systemic acquired resistance
- PMID: 22147520
- PMCID: PMC3271772
- DOI: 10.1104/pp.111.187468
Next-generation systemic acquired resistance
Abstract
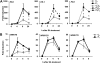
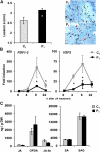
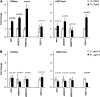
Systemic acquired resistance (SAR) is a plant immune response to pathogen attack. Recent evidence suggests that plant immunity involves regulation by chromatin remodeling and DNA methylation. We investigated whether SAR can be inherited epigenetically following disease pressure by Pseudomonas syringae pv tomato DC3000 (PstDC3000). Compared to progeny from control-treated Arabidopsis (Arabidopsis thaliana; C(1)), progeny from PstDC3000-inoculated Arabidopsis (P(1)) were primed to activate salicylic acid (SA)-inducible defense genes and were more resistant to the (hemi)biotrophic pathogens Hyaloperonospora arabidopsidis and PstDC3000. This transgenerational SAR was sustained over one stress-free generation, indicating an epigenetic basis of the phenomenon. Furthermore, P(1) progeny displayed reduced responsiveness of jasmonic acid (JA)-inducible genes and enhanced susceptibility to the necrotrophic fungus Alternaria brassicicola. This shift in SA- and JA-dependent gene responsiveness was not associated with changes in corresponding hormone levels. Instead, chromatin immunoprecipitation analyses revealed that SA-inducible promoters of PATHOGENESIS-RELATED GENE1, WRKY6, and WRKY53 in P(1) plants are enriched with acetylated histone H3 at lysine 9, a chromatin mark associated with a permissive state of transcription. Conversely, the JA-inducible promoter of PLANT DEFENSIN1.2 showed increased H3 triple methylation at lysine 27, a mark related to repressed gene transcription. P(1) progeny from the defense regulatory mutant non expressor of PR1 (npr1)-1 failed to develop transgenerational defense phenotypes, demonstrating a critical role for NPR1 in expression of transgenerational SAR. Furthermore, the drm1drm2cmt3 mutant that is affected in non-CpG DNA methylation mimicked the transgenerational SAR phenotype. Since PstDC3000 induces DNA hypomethylation in Arabidopsis, our results suggest that transgenerational SAR is transmitted by hypomethylated genes that direct priming of SA-dependent defenses in the following generations.
Figures


Comment in
-
Prime time for transgenerational defense.Plant Physiol. 2012 Feb;158(2):545. doi: 10.1104/pp.112.900430. Plant Physiol. 2012. PMID: 22308198 Free PMC article. No abstract available.
References
-
- Baulcombe D, Crute I, Davies B, Dunwell J, Gale M, Jones J, Pretty J, Sutherland W, Toumin C. (2009) Reaping the benefits: science and the sustainable intensification of global agriculture. In Royal Society, RS Policy Document 11/09. Royal Society, London, 2009
-
- Brodersen P, Voinnet O. (2006) The diversity of RNA silencing pathways in plants. Trends Genet 22: 268–280 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

