OsHsfA2c and OsHsfB4b are involved in the transcriptional regulation of cytoplasmic OsClpB (Hsp100) gene in rice (Oryza sativa L.)
- PMID: 22147560
- PMCID: PMC3273560
- DOI: 10.1007/s12192-011-0303-5
OsHsfA2c and OsHsfB4b are involved in the transcriptional regulation of cytoplasmic OsClpB (Hsp100) gene in rice (Oryza sativa L.)
Abstract
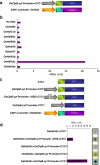
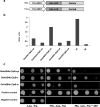
ClpB-cytoplasmic (ClpB-cyt)/Hsp100 is an important chaperone protein in rice. Cellular expression of OsClpB-cyt transcript is governed by heat stress, metal stress, and developmental cues. Transgenic rice plants produced with 2 kb OsClpB-cyt promoter driving Gus reporter gene showed heat- and metal-regulated Gus expression in vegetative tissues and constitutive Gus expression in calli, flowering tissues, and embryonal half of seeds. Rice seedlings regenerated with OsClpB-cyt promoter fragment with deletion of its canonical heat shock element sequence (HSE(-273 to -280)) showed not only heat shock inducibility of Gus transcript/protein but also constitutive expression of Gus in vegetative tissues. It thus emerges that the only classical HSE present in OsClpB-cyt promoter is involved in repressing expression of OsClpB-cyt transcript under unstressed control conditions. Yeast one-hybrid assays suggested that OsHsfA2c specifically interacts with OsClpB-cyt promoter. OsHsfA2c also showed binding with OsClpB-cyt and OsHsfB4b showed binding with OsClpB-cyt; notably, interaction of OsHsfB4b was seen for all three OsClpB/Hsp100 protein isoforms (i.e., ClpB-cytoplasmic, ClpB-mitochondrial, and ClpB-chloroplastic). Furthermore, OsHsfB4b showed interaction with OsHsfA2c. This study suggests that OsHsfA2c may play a role as transcriptional activator and that OsHsfB4b is an important part of this heat shock responsive circuitry.
Figures
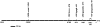



Similar articles
-
Characterization of 5'UTR of rice ClpB-C/Hsp100 gene: evidence of its involvement in post-transcriptional regulation.Cell Stress Chaperones. 2016 Mar;21(2):271-83. doi: 10.1007/s12192-015-0657-1. Epub 2015 Nov 6. Cell Stress Chaperones. 2016. PMID: 26546418 Free PMC article.
-
Genome-wide analysis of rice ClpB/HSP100, ClpC and ClpD genes.BMC Genomics. 2010 Feb 8;11:95. doi: 10.1186/1471-2164-11-95. BMC Genomics. 2010. PMID: 20141629 Free PMC article.
-
Cycloheximide-mediated superinduction of genes involves both native and foreign transcripts in rice (Oryza sativa L.).Plant Physiol Biochem. 2011 Jan;49(1):9-12. doi: 10.1016/j.plaphy.2010.09.010. Epub 2010 Oct 1. Plant Physiol Biochem. 2011. PMID: 20980158
-
Binding affinities and interactions among different heat shock element types and heat shock factors in rice (Oryza sativa L.).FEBS J. 2011 Sep;278(17):3076-85. doi: 10.1111/j.1742-4658.2011.08229.x. Epub 2011 Aug 2. FEBS J. 2011. PMID: 21729241
-
ClpB/Hsp100 proteins and heat stress tolerance in plants.Crit Rev Biotechnol. 2016 Oct;36(5):862-74. doi: 10.3109/07388551.2015.1051942. Epub 2015 Jun 30. Crit Rev Biotechnol. 2016. PMID: 26121931 Review.
Cited by
-
OsHsfB4b Confers Enhanced Drought Tolerance in Transgenic Arabidopsis and Rice.Int J Mol Sci. 2022 Sep 16;23(18):10830. doi: 10.3390/ijms231810830. Int J Mol Sci. 2022. PMID: 36142741 Free PMC article.
-
Heat shock transcription factor 3 regulates plant immune response through modulation of salicylic acid accumulation and signalling in cassava.Mol Plant Pathol. 2018 Oct;19(10):2209-2220. doi: 10.1111/mpp.12691. Epub 2018 Jul 19. Mol Plant Pathol. 2018. PMID: 29660238 Free PMC article.
-
Characterization of 5'UTR of rice ClpB-C/Hsp100 gene: evidence of its involvement in post-transcriptional regulation.Cell Stress Chaperones. 2016 Mar;21(2):271-83. doi: 10.1007/s12192-015-0657-1. Epub 2015 Nov 6. Cell Stress Chaperones. 2016. PMID: 26546418 Free PMC article.
-
Modification of Serine 1040 of SIBRI1 Increases Fruit Yield by Enhancing Tolerance to Heat Stress in Tomato.Int J Mol Sci. 2020 Oct 16;21(20):7681. doi: 10.3390/ijms21207681. Int J Mol Sci. 2020. PMID: 33081382 Free PMC article.
-
A seed preferential heat shock transcription factor from wheat provides abiotic stress tolerance and yield enhancement in transgenic Arabidopsis under heat stress environment.PLoS One. 2013 Nov 12;8(11):e79577. doi: 10.1371/journal.pone.0079577. eCollection 2013. PLoS One. 2013. PMID: 24265778 Free PMC article.
References
-
- Agarwal M, Sahi C, Katiyar-Agarwal S, Agarwal S, Young T, Gallie DR, Sharma VM, Ganesan K, Grover A. Molecular characterization of rice hsp101: complementation of yeast hsp104 mutation by disaggregation of protein granules and differential expression in indica and japonica rice types. Plant Mol Biol. 2003;51:543–553. doi: 10.1023/A:1022324920316. - DOI - PubMed
-
- Baniwal SK, Bharti K, Chan KY, Fauth M, Ganguli A, Kotak S, Mishra SK, Nover L, Port M, Scharf KD, Tripp J, Weber C, Zielinski D, Koskull-Döring P. Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J Biosci. 2004;29:471–487. doi: 10.1007/BF02712120. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

