Global gene expression profiling in mouse plasma cell tumor precursor and bystander cells reveals potential intervention targets for plasma cell neoplasia
- PMID: 22147894
- PMCID: PMC3271714
- DOI: 10.1182/blood-2011-06-363887
Global gene expression profiling in mouse plasma cell tumor precursor and bystander cells reveals potential intervention targets for plasma cell neoplasia
Abstract
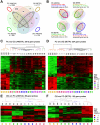
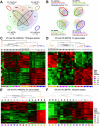
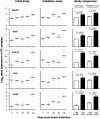
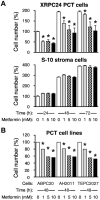
Tumor progression usually proceeds through several sequential stages, any of which could be targets for interrupting the progression process if one understood these steps at the molecular level. We extracted nascent plasma cell tumor (PCT) cells from within inflammatory oil granulomas (OG) isolated from IP pristane-injected BALB/c.iMyc(Eμ) mice at 5 different time points during tumor progression. We used laser capture microdissection to collect incipient PCT cells and analyzed their global gene expression on Affymetrix Mouse Genome 430A microarrays. Two independent studies were performed with different sets of mice. Analysis of the expression data used ANOVA and Bayesian estimation of temporal regulation. Genetic pathway analysis was performed using MetaCore (GeneGo) and IPA (Ingenuity). The gene expression profiles of PCT samples and those of undissected OG samples from adjacent sections showed that different genes and pathways were mobilized in the tumor cells during tumor progression, compared with their stroma. Our analysis implicated several genetic pathways in PCT progression, including biphasic (up- and then down-regulation) of the Spp1/osteopontin-dependent network and up-regulation of mRNA translation/protein synthesis. The latter led to a biologic validation study that showed that the AMPK-activating diabetes drug, metformin, was a potent specific PCT inhibitor in vitro.
Figures


Similar articles
-
Gene expression profiling reveals different pathways related to Abl and other genes that cooperate with c-Myc in a model of plasma cell neoplasia.BMC Genomics. 2007 Aug 31;8:302. doi: 10.1186/1471-2164-8-302. BMC Genomics. 2007. PMID: 17764563 Free PMC article.
-
Molecular and cytological features of the mouse B-cell lymphoma line iMycEmu-1.Mol Cancer. 2005 Nov 9;4:40. doi: 10.1186/1476-4598-4-40. Mol Cancer. 2005. PMID: 16277667 Free PMC article.
-
CDDO-Imidazolide inhibits growth and survival of c-Myc-induced mouse B cell and plasma cell neoplasms.Mol Cancer. 2006 Jun 7;5:22. doi: 10.1186/1476-4598-5-22. Mol Cancer. 2006. PMID: 16759389 Free PMC article.
-
Analysis of gene expression in prostate cancer epithelial and interstitial stromal cells using laser capture microdissection.BMC Cancer. 2010 Apr 28;10:165. doi: 10.1186/1471-2407-10-165. BMC Cancer. 2010. PMID: 20426842 Free PMC article.
-
Cyclooxygenase-2 inhibitors down-regulate osteopontin and Nr4A2-new therapeutic targets for colorectal cancers.Gastroenterology. 2009 Oct;137(4):1358-66.e1-3. doi: 10.1053/j.gastro.2009.06.039. Epub 2009 Jun 21. Gastroenterology. 2009. PMID: 19549529
Cited by
-
(18)F-FDG-PET/CT imaging in an IL-6- and MYC-driven mouse model of human multiple myeloma affords objective evaluation of plasma cell tumor progression and therapeutic response to the proteasome inhibitor ixazomib.Blood Cancer J. 2013 Nov 29;3(11):e165. doi: 10.1038/bcj.2013.61. Blood Cancer J. 2013. PMID: 24292417 Free PMC article.
-
Impact of metformin on the progression of MGUS to multiple myeloma.Leuk Lymphoma. 2017 May;58(5):1265-1267. doi: 10.1080/10428194.2016.1236375. Epub 2016 Oct 5. Leuk Lymphoma. 2017. PMID: 27701912 Free PMC article. No abstract available.
-
Discovering Potential in Non-Cancer Medications: A Promising Breakthrough for Multiple Myeloma Patients.Cancers (Basel). 2024 Jun 28;16(13):2381. doi: 10.3390/cancers16132381. Cancers (Basel). 2024. PMID: 39001443 Free PMC article. Review.
-
Transgenic mouse model of IgM+ lymphoproliferative disease mimicking Waldenström macroglobulinemia.Blood Cancer J. 2016 Nov 4;6(11):e488. doi: 10.1038/bcj.2016.95. Blood Cancer J. 2016. PMID: 27813533 Free PMC article.
-
Association between Leptin and Complement in Hepatitis C Patients with Viral Clearance: Homeostasis of Metabolism and Immunity.PLoS One. 2016 Nov 21;11(11):e0166712. doi: 10.1371/journal.pone.0166712. eCollection 2016. PLoS One. 2016. PMID: 27870883 Free PMC article.
References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. - PubMed
-
- Davies FE, Dring AM, Li C, et al. Insights into the multistep transformation of MGUS to myeloma using microarray expression analysis. Blood. 2003;102(13):4504–4511. - PubMed
-
- Zhan F, Hardin J, Kordsmeier B, et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood. 2002;99(5):1745–1757. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

