Trinucleotide repeat DNA alters structure to minimize the thermodynamic impact of 8-oxo-7,8-dihydroguanine
- PMID: 22148399
- PMCID: PMC3254800
- DOI: 10.1021/bi201552s
Trinucleotide repeat DNA alters structure to minimize the thermodynamic impact of 8-oxo-7,8-dihydroguanine
Abstract
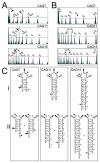
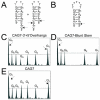
In the phenomenon of trinucleotide repeat (TNR) expansion, an important interplay exists between DNA damage repair of 8-oxo-7,8-dihydroguanine (8-oxoG) and noncanonical structure formation. We show that TNR DNA adapts its structure to accommodate 8-oxoG. Using chemical probe analysis, we find that CAG repeats composing the stem-loop arm of a three-way junction alter the population of structures in response to 8-oxoG by positioning the lesion at or near the loop. Furthermore, we find that oligonucleotides composed of odd-numbered repeat sequences, which form populations of two structures, will also alter their structure to place 8-oxoG in the loop. However, sequences with an even number of repeats do not display this behavior. Analysis by differential scanning calorimetry indicates that when the lesion is located within the loop, there are no significant changes to the thermodynamic parameters as compared to the DNA lacking 8-oxoG. This contrasts with the enthalpic destabilization observed when 8-oxoG is base-paired to C and indicates that positioning 8-oxoG in the loop avoids the thermodynamic penalty associated with 8-oxoG base-pairing. Since formation of stem-loop hairpins is proposed to facilitate TNR expansion, these results highlight the importance of defining the structural consequences of DNA damage.
Figures




Similar articles
-
Incidence and persistence of 8-oxo-7,8-dihydroguanine within a hairpin intermediate exacerbates a toxic oxidation cycle associated with trinucleotide repeat expansion.DNA Repair (Amst). 2011 Aug 15;10(8):887-96. doi: 10.1016/j.dnarep.2011.06.003. Epub 2011 Jul 2. DNA Repair (Amst). 2011. PMID: 21727036 Free PMC article.
-
Structure-dependent DNA damage and repair in a trinucleotide repeat sequence.Biochemistry. 2009 Jul 21;48(28):6655-63. doi: 10.1021/bi9007403. Biochemistry. 2009. PMID: 19527055
-
Unique Length-Dependent Biophysical Properties of Repetitive DNA.J Phys Chem B. 2016 May 12;120(18):4195-203. doi: 10.1021/acs.jpcb.6b00927. Epub 2016 May 3. J Phys Chem B. 2016. PMID: 27115707 Free PMC article.
-
DNA base excision repair: a mechanism of trinucleotide repeat expansion.Trends Biochem Sci. 2012 Apr;37(4):162-72. doi: 10.1016/j.tibs.2011.12.002. Epub 2012 Jan 27. Trends Biochem Sci. 2012. PMID: 22285516 Free PMC article. Review.
-
The central role of DNA damage and repair in CAG repeat diseases.Dis Model Mech. 2018 Jan 30;11(1):dmm031930. doi: 10.1242/dmm.031930. Dis Model Mech. 2018. PMID: 29419417 Free PMC article. Review.
Cited by
-
Frustration Between Preferred States of Complementary Trinucleotide Repeat DNA Hairpins Anticorrelates with Expansion Disease Propensity.J Mol Biol. 2023 May 15;435(10):168086. doi: 10.1016/j.jmb.2023.168086. Epub 2023 Apr 5. J Mol Biol. 2023. PMID: 37024008 Free PMC article.
-
The degree of global DNA hypomethylation in peripheral blood correlates with that in matched tumor tissues in several neoplasia.PLoS One. 2014 Mar 20;9(3):e92599. doi: 10.1371/journal.pone.0092599. eCollection 2014. PLoS One. 2014. PMID: 24651295 Free PMC article.
-
An oxidized abasic lesion inhibits base excision repair leading to DNA strand breaks in a trinucleotide repeat tract.PLoS One. 2018 Feb 1;13(2):e0192148. doi: 10.1371/journal.pone.0192148. eCollection 2018. PLoS One. 2018. PMID: 29389977 Free PMC article.
-
Dynamic DNA Energy Landscapes and Substrate Complexity in Triplet Repeat Expansion and DNA Repair.Biomolecules. 2019 Nov 6;9(11):709. doi: 10.3390/biom9110709. Biomolecules. 2019. PMID: 31698848 Free PMC article.
-
Base excision repair of oxidative DNA damage coupled with removal of a CAG repeat hairpin attenuates trinucleotide repeat expansion.Nucleic Acids Res. 2014 Apr;42(6):3675-91. doi: 10.1093/nar/gkt1372. Epub 2014 Jan 14. Nucleic Acids Res. 2014. PMID: 24423876 Free PMC article.
References
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. - PubMed
-
- Steenken S, Jovanovic S. How Easily Oxidizable Is DNA? One-Electron Reduction Potentials of Adenosine and Guanosine Radicals in Aqueous Solution. J. Am. Chem. Soc. 1997;119:617–618.
-
- Burrows CJ, Muller JG. Oxidative Nucleobase Modifications Leading to Strand Scission. Chem. Rev. 1998;98:1109–1152. - PubMed
-
- David SS, Williams SD. Chemistry of Glycosylases and Endonucleases Involved in Base-Excision Repair. Chem. Rev. 1998;98:1221–1262. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

